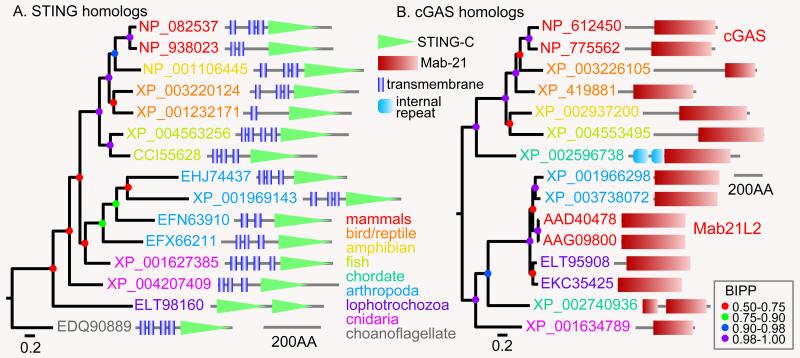
Figure 1. Phylogenetic analysis of STING and cGAS proteins.
The major vertebrate and invertebrate phyla were individually screened for homologs of human STING and cGAS proteins using BlastP. A few representative best hits for each phylum were selected, which were then used for a reverse screen of mammalian genomes. For STING, the reverse search yielded mouse and bat STING proteins. However, invertebrate cGAS homologs yielded mammalian Mab21 proteins as bidirectional hits. Protein sequences were aligned using M-coffee (53). After deletion of segments with poor consensus alignment, sequences were subjected to Bayesian inference for establishment of phylogenetic relationships between proteins (54). Analysis were run for 1 million generations under a mixed amino-acid model with rate variation between sites estimated by a gamma distribution. Bayesian inference posterior probabilities (BIPPs) of tree nodes are indicated by colored dots. Gene identifiers of the proteins are annotated with functional domain architectures and color-coded to represent the phyla from which they are derived. Corresponding species names are listed in the legends to supplemental figures 1 and 2.