Table 2. Comparison of search results for cell (80.36, 80.36, 99.44, 90, 90, 120) from entry 1u4j in space group R3 (see Le Trong Stenkamp, 2007 ▶).
In each case the PDB code (Bernstein etal., 1977 ▶; Berman etal., 2000 ▶) found is shown with the distance metric for that method. In the case of Nearest-Cell (Ramraj etal., 2011 ▶), a second column with the square root of the metric is provided as well. The results are sorted by the NCDist distance. Results have been cut off at 3.5 in the NCDist metric. The three alternative cells cited by Le Trong Stenkamp (2007 ▶) are in bold and marked with an asterisk (*). The Nearest-Cell (‘N-C’) results are from the http://www.strubi.ox.ac.uk/nearest-cell/nearest-cell.cgi web site. The V7, NCDist (‘NCD’), 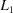 and
and 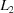 results are from SAUC.
results are from SAUC.
| PDB ID | N-C | 3(N-C)1/2 | NCD | V7 | L 1 | L 2 | Molecule | EC code |
|---|---|---|---|---|---|---|---|---|
| 1u4j* | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | Phospholipase A2 isoform 2 | 3.1.1.4 |
| 1g0z | 0 | 0 | 0 | 0 | 0 | 0 | Phospholipase A2 | 3.1.1.4 |
| 1g2x* | 0.1 | 1.0 | 0.9 | 0.2 | 0.4 | 0.5 | Phospholipase A2 | 3.1.1.4 |
| 2osn | 0.9 | 0.2 | 0.4 | 0.5 | Phospholipase A2 isoform 3 | 3.1.1.4 | ||
| 2cmp | 0.3 | 1.7 | 1.5 | 0.7 | 1.3 | 1.5 | Terminase small subunit | |
| 3kp8 | 0.43 | 0.66 | 1.1 | 1.7 | 1.5 | 1.8 | VKORC1/thioredoxin domain protein | |
| 3mij | 1.7 | 1.0 | 0.8 | 0.8 | RNA [5-R(*UP*AP*GP*GP*GP*UP*UP*AP*GP*GP*GP*U)-3] | |||
| 3e56 | 0.4 | 1.9 | 1.9 | 1.6 | 1.5 | 1.5 | Putative uncharacterized protein | |
| 1csq | 0.5 | 2.1 | 2.0 | 1.8 | 1.2 | 1.6 | Cold shock protein B (CSPB) | |
| 4den | 2.1 | 2.0 | 2.0 | 2.0 | Actinohivin | |||
| 3svi | 0.5 | 2.2 | 2.1 | 1.9 | 1.5 | 1.8 | Type III effector HopAB2 | |
| 1fkf | 0.8 | 2.7 | 2.4 | 2.7 | 2.4 | 2.9 | FK506 binding protein | 5.2.1.8 |
| 1fkj | 0.8 | 2.7 | 2.4 | 2.7 | 2.4 | 2.9 | FK506 binding protein | 5.2.1.8 |
| 1fkd | 0.9 | 2.8 | 2.5 | 2.8 | 2.5 | 3.0 | FK506 binding protein | 5.2.1.8 |
| 1bkf | 0.9 | 2.9 | 2.5 | 2.8 | 3.1 | 3.4 | Subtilisin Carlsberg | 3.4.21.62 |
| 2fke | 0.9 | 2.9 | 2.6 | 3.0 | 2.5 | 3.1 | FK506 binding protein | 5.2.1.8 |
| 3tjy | 0.9 | 2.8 | 2.6 | 3.0 | 2.5 | 2.9 | Effector protein HopAB3 | |
| 2i5l | 1.1 | 3.1 | 2.7 | 3.7 | 4.3 | 4.4 | Cold shock protein CSPB | |
| 3p63 | 1.3 | 3.4 | 2.9 | 4.0 | 4.4 | 4.9 | Ferredoxin | |
| 2wce | 1.2 | 3.3 | 3.0 | 3.5 | 5.9 | 6.4 | Protein S100-A12 | |
| 4sga | 1.4 | 3.5 | 3.1 | 4.8 | 5.7 | 5.8 | Proteinase A (SGPA) | 3.4.21.80 |
| 2cxd | 1.4 | 3.5 | 3.1 | 4.8 | 4.7 | 5.4 | Conserved hypothetical protein, TTHA0068 | |
| 5sga | 1.4 | 3.5 | 3.1 | 4.9 | 5.8 | 5.9 | Proteinase A (SGPA) | 3.4.21.80 |
| 2sga | 1.4 | 3.6 | 3.1 | 4.9 | 5.8 | 5.9 | Proteinase A | 3.4.21.80 |
| 3sga | 1.4 | 3.6 | 3.1 | 5.0 | 5.9 | 6.0 | Proteinase A (SGPA) | 3.4.21.80 |
| 1f9p | 1.4 | 3.5 | 3.1 | 4.7 | 4.1 | 4.8 | Connective tissue activating peptide III | |
| 2yzu | 3.1 | 4.8 | 6.3 | 6.7 | Thioredoxin | |||
| 1sgc | 1.5 | 3.7 | 3.2 | 5.2 | 6.2 | 6.3 | Proteinase A | 3.4.21.80 |
| 1pkr | 1.4 | 3.6 | 3.2 | 4.7 | 3.9 | 4.9 | Plasminogen | 3.4.21.7 |
| 1gus | 1.2 | 3.2 | 3.2 | 0.3 | 3.4 | 4.6 | Molybdate binding protein II | |
| 2vri | 1.5 | 3.7 | 3.2 | 5.2 | 5.0 | 5.7 | Non-structural protein 3 | 3.4.19.12 |
| 2cvk | 3.2 | 5.2 | 7.0 | 7.3 | Thioredoxin | |||
| 2c9q | 1.5 | 3.6 | 3.3 | 4.8 | 4.6 | 4.9 | Copper resistance protein C | |
| 1fe5* | 1.2 | 3.3 | 3.3 | 2.3 | 6.0 | 7.0 | Phospholipase A2 | 3.1.1.4 |
| 1dpy | 1.2 | 3.3 | 3.3 | 2.3 | 6.0 | 7.0 | Phospholipase A2 | 3.1.1.4 |
| 2he2 | 1.5 | 3.6 | 3.3 | 4.8 | 4.5 | 5.1 | Discs large homolog 2 | |
| 3su6 | 1.5 | 3.7 | 3.3 | 5.0 | 4.5 | 5.4 | NS3 protease, NS4A protein | |
| 1cdc | 3.3 | 4.9 | 4.3 | 5.4 | CD2 | |||
| 1gut | 1.2 | 3.3 | 3.3 | 0.1 | 3.4 | 4.8 | Molybdate binding protein II | |
| 2it5 | 1.6 | 3.8 | 3.3 | 5.6 | 5.7 | 6.4 | CD209 antigen, DCSIGN-CRD | |
| 3su5 | 1.6 | 3.8 | 3.3 | 5.1 | 4.8 | 5.7 | NS3 protease, NS4A protein | |
| 3su1 | 1.6 | 3.8 | 3.3 | 5.2 | 4.7 | 5.7 | Genome polyprotein | |
| 3su2 | 1.6 | 3.8 | 3.3 | 5.2 | 4.7 | 5.6 | Genome polyprotein | |
| 3cyo | 3.4 | 5.6 | 7.6 | 7.9 | Transmembrane protein | |||
| 1sl4 | 1.7 | 3.9 | 3.4 | 5.8 | 6.2 | 6.8 | mDC-SIGN1B type I isoform | |
| 3su3 | 1.6 | 3.8 | 3.4 | 5.3 | 4.9 | 5.9 | NS3 protease, NS4A protein | |
| 2it6 | 1.7 | 3.9 | 3.4 | 6.0 | 6.4 | 7.0 | CD209 antigen | |
| 3sv7 | 1.7 | 3.9 | 3.5 | 5.6 | 5.5 | 6.4 | NS3 protease, NS4A protein |
