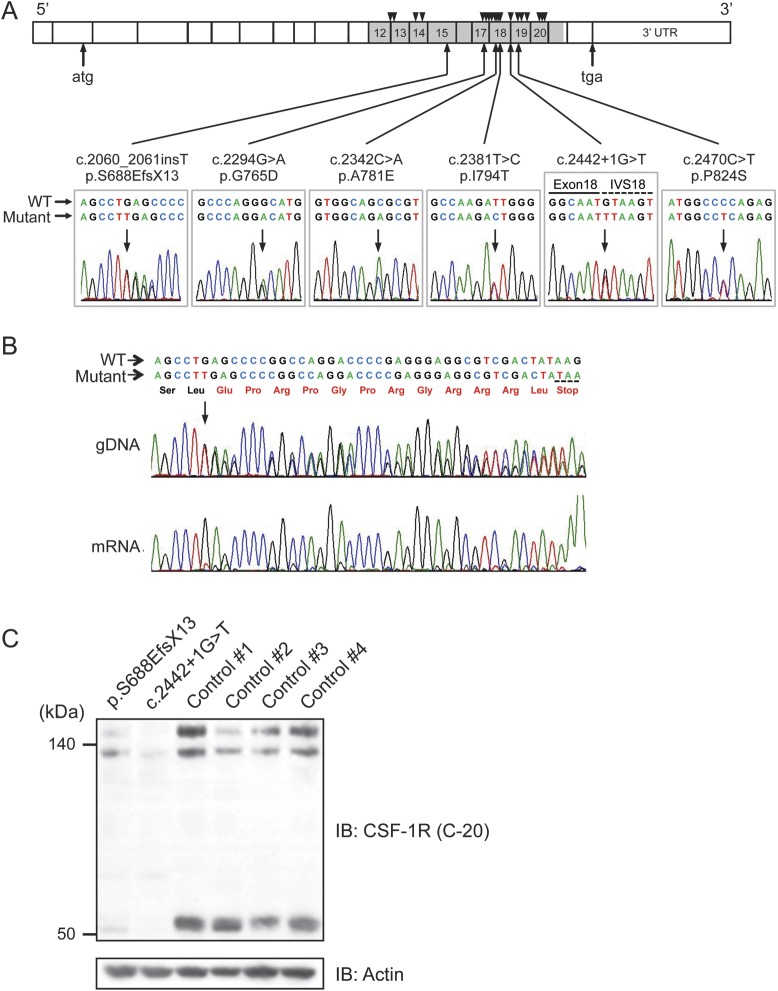
Figure 1. Identification of CSF-1R mutations, and mRNA and protein expressions of mutant CSF-1R.
(A) Schematic illustration of CSF-1R structure. Six different mutations identified in this study are shown below the diagram of electropherograms. The tyrosine kinase domain of CSF-1R is shown in gray. Numbers represent exons in which mutations were identified. Positions of previously reported mutations are shown as triangles above the diagram. IVS = intervening sequence. (B) Sequencing electropherogram of amplified genomic DNA and reverse transcription PCR amplicons of patient with frameshift mutation (p.S688EfsX13), which was predicted to undergo nonsense-mediated mRNA decay. The expression of the mutant allele was hardly detectable, suggesting that this frameshift mutation results in nonsense-mediated decay of mutant mRNA. The predicted amino acid sequences followed by a stop codon are shown in red. (C) Immunoblot analysis of CSF-1R protein using anti-CSF-1R antibody (C-20) in Triton X-100 soluble fraction from frontal cortex of autopsied cases (c.2442+1G>T and p.S688EfsX13) and 4 control subjects (1–4) without neurologic disorder. Note that the full-length CSF-1R (150 kDa and 130 kDa, representing mature protein and immature full-length protein, respectively) and proteolytically cleaved C-terminal fragment of CSF-1R (55 kDa) showed markedly decreased expression levels. The equivalency of protein loading is shown in the actin blot (bottom).