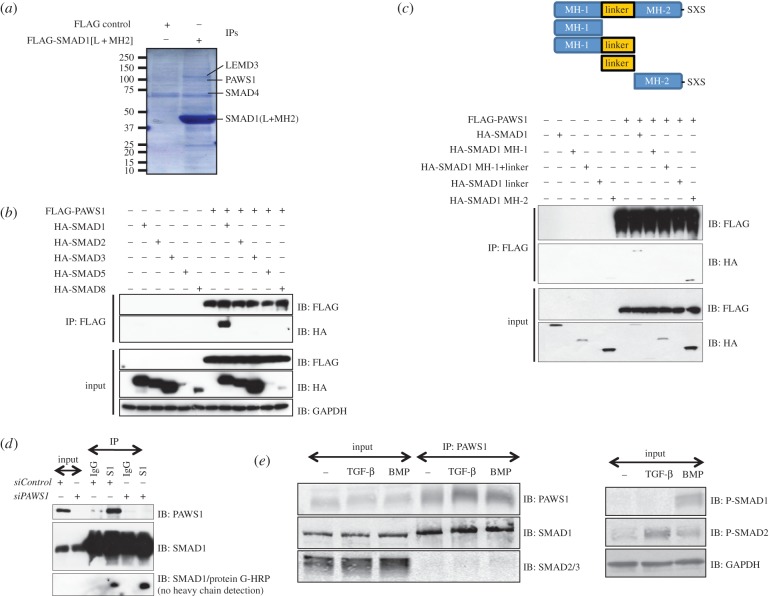
Figure 1.
PAWS1 interacts with SMAD1. (a) Anti-FLAG IPs from HEK293 extracts transfected with vectors either encoding FLAG control or FLAG-tagged SMAD1[L + MH2] fragment were incubated with HeLa extracts. Elution was performed with 3X FLAG peptide. Eluted proteins were denatured and resolved by SDS–PAGE and the gel was Coomassie-stained. Gel pieces (2 mm) covering the entire lane of each sample were excised for identification by mass fingerprinting. The positions of some of the identified proteins are indicated. (b) HEK293 cells were transfected with the indicated HA-SMAD constructs either alone or together with FLAG-PAWS1 construct. The extracts and anti-FLAG IPs were analysed by immunoblotting using the indicated antibodies. (c) HEK293 cells transfected with constructs encoding either HA-SMAD1 or indicated HA-SMAD1 truncation mutants either individually or together with construct encoding FLAG-PAWS1. The extracts and anti-FLAG IPs were analysed by immunoblotting using the indicated antibodies. (d) HaCaT cells were transfected with a pool of two different siRNAs against either PAWS1 (150 pM each), or with siRNA against FOXO4, for 48 h prior to lysis. Extracts and IPs, using either anti-SMAD1 antibody or pre-immune IgG, were analysed by immunoblotting using the indicated antibodies. For SMAD1/protein-G-HRP immunoblot, the membrane was first blocked in 5% milk containing 500 ng ml−1 protein G, incubated with SMAD1 antibody as primary, and protein-G HRP was used as secondary. This strategy excludes the detection of antibody heavy chains in IP samples. (e) HaCaT cells were treated with either BMP-2 (25 ng ml−1; 1 h), TGF-β (50 pM; 1 h) or left untreated prior to lysis. Extracts and anti-PAWS1 IPs were analysed by immunoblotting with the indicated antibodies.