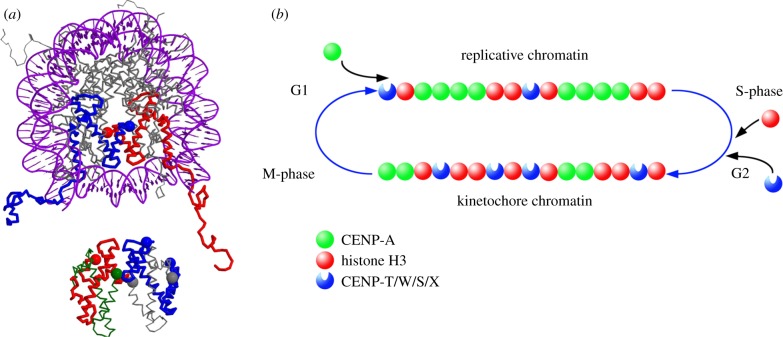
Figure 6.
Model of CENP-T/W/S/X in chromatin. (a) Potential position of the CENP-TWSX tetramer relative to a classical nucleosome. Top: top view of the nucleosomal structure including DNA (1kx5, [49]) with both H3 molecules coloured in red and blue, respectively; the H3 C-termini are indicated by coloured spheres. Bottom: structure of the CENP-TWSX tetramer (3vh5; [30]) positioned in the plane relative to the nucleosomal structure. The tetramer is assumed to be partly surrounded by DNA, and accordingly, space is left between the two structures (2 nm). Colour coding: CENP-T, red with C-terminus indicated by a large sphere; CENP-W, green with C-terminus indicated by a large sphere; CENP-S, blue with both termini marked with a large sphere; CENP-X, grey with both termini marked with a large sphere. Only well-determined termini positions are shown. The largest distance of 8.4 nm is found between the H3 C-terminus and the CENP-X N-terminus, thus all termini shown are sufficiently proximal for FRET. Note the close proximities of the CENP-S and -X termini, explaining well the measured FRET. Shown is one potential positioning of the two complexes, also alternative arrangements (for example, the CENP-TWSX tetramer between two classical nucleosomes) would explain well the measured FRET results. (b) Proposed switching mechanism for converting centromeric chromatin between functional states. Completion of CENP-A assembly results in replication-competent chromatin, top, which is segregated to daughter chromatids during S phase. Assembly of CENP-T/W/S/X, probably in conjunction with histone H3, is activated by DNA replication, resulting in kinetochore-competent chromatin, bottom, with a distinctive subunit composition for function in mitosis.