Figure 6.
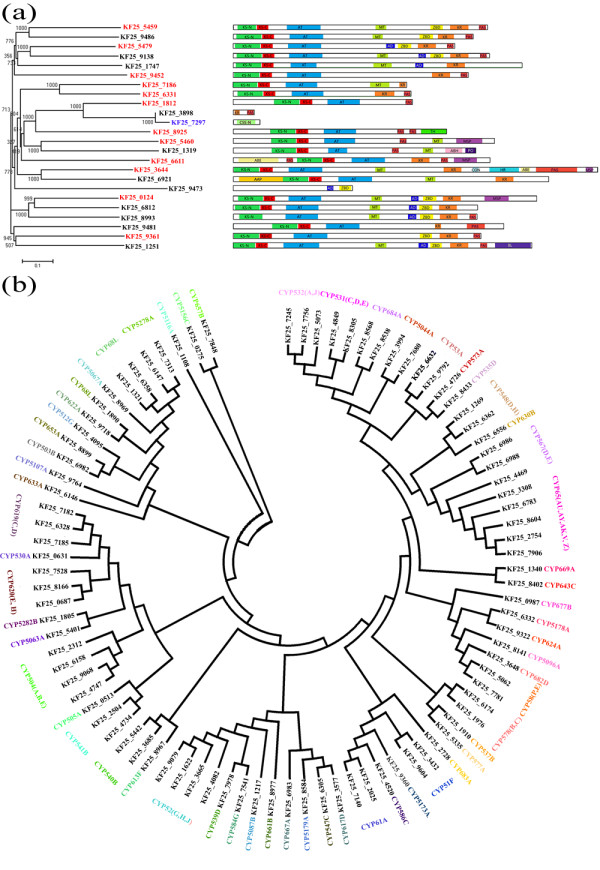
Neighbor-joining phylogenetic tree of polyketide synthases (PKSs) and cytochrome P450 from the KF-25 genome. The amino acid sequences of the proteins were used to construct the phylogenetic tree using ClustalX2.0 with the neighbor-joining method. The branch length scale bar below the phylogenetic tree indicates the number of substitutions per amino acid site. (a) The functional domain architecture of proteins was predicted using Pfam and AMSPKS [89,90] and is shown on the right. Protein domain names were as follows: KS_N, β-ketoacyl synthase, N-terminal domain; KS_C, β-ketoacyl synthase, N-terminal domain; AT, acyl transferase; KR, β-keto reductase; PAS, phosphopantetheine attachment site; MT, methyltransferase; MSP, male sterility protein; TH, thioesterase; ZBD, zinc-binding dehydrogenase; ER, ER domain; AD, alcohol dehydrogenase GroES-like domain; CSS_N, chalcone and stilbene synthases, N-terminal domain; ABH, α/βhydrolase; PO, prolyl oligopeptidase; CON, condensation domain; HR, HxxPF-repeated domain; ABE, AMP-binding enzyme; BL, β-lactamase; AAP, amino acid permease. The ORFs indicated in red (type I PKS) and blue (type II PKS) are the members of the putative secondary metabolism pathways. (b) The cytochrome P450 (CYPs) identified in the KF-25 genome and CYPs of different families are indicated in different colors. The families of the corresponding CYPs are indicated beside the name of the proteins.
