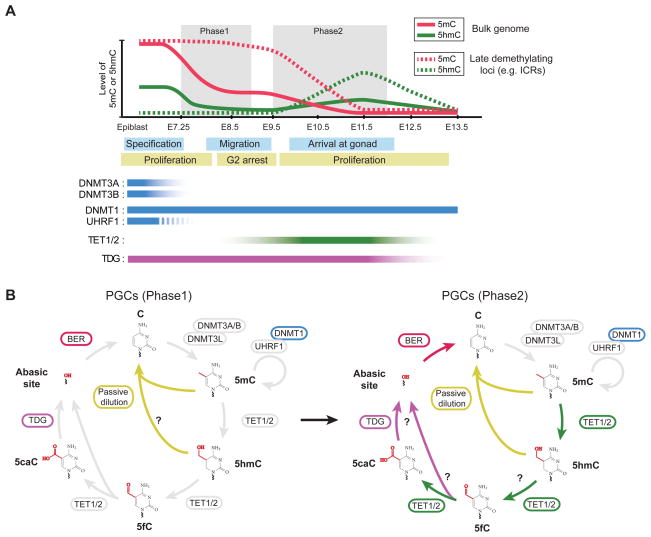
Figure 5. Global reprogramming of DNA methylation patterns during early PGC development.
(A) Dynamic changes in cytosine modifications and expression levels of relevant enzymes during early stage of mouse PGC development. Global erasure of DNA methylation in developing PGCs goes through two phases. In the first phase (E7.25 to E9.0), both de novo and maintenance methylation machineries are functionally impaired due to the repression of DNMT3A/3B/3L and UHRF1. There is also evidence that remaining UHRF1 is excluded from nucleus (dash line). Thus, loss of bulk genomic DNA methylation (solid line, inherited from epiblast cell stage) takes place in a replication-dependent manner in phase 1. However, a portion of the genome (dash line) including ICRs and germline-specific genes are partially or fully protected from the first wave of passive demethylation. In the second phase (E9.5 to E12.0), TET1 (possibly TET2) are up-regulated and oxidize remaining 5mC to 5hmC.
(B) Developmental stage-specific usage of the cytosine modifying enzymatic cascade during early stage of PGC development. In phase 1 of PGC demethylation, bulk of the genome undergoes global demethylation in the absence of de novo and maintenance DNA methylation. In phase 2, TET1/2 proteins oxidize remaining 5mC at late demethylating loci. Replication-dependent loss of 5hmC completes the global demethylation process. 5fC/5caC is not specifically enriched in developing PGCs as compared to surrounding somatic cells, so they may be actively excised by TDG/BER pathway that is present in PGCs.