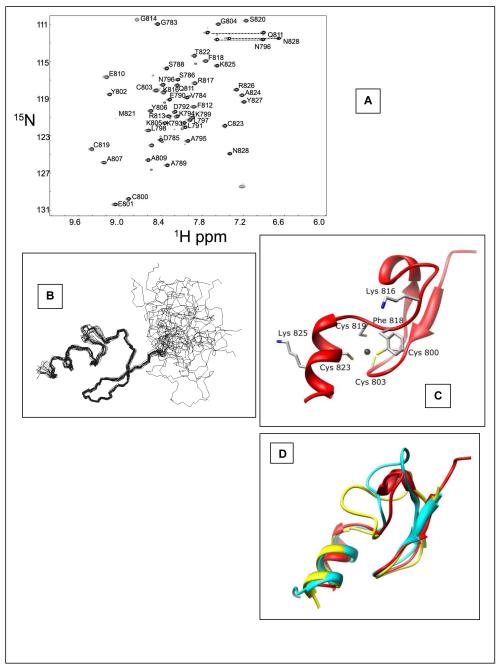
Figure 3.
Solution structure determination of the hPh1 FCS domain. A. hPh1 FCS {1H}-15N HSQC. The chemical shift assignment for each residue is indicated. B. Ensemble of the 20 lowest energy structures of the hPh1 FCS domain. The RMSD of secondary structure backbone is 0.46. C. Ribbon diagram of the hPh1 FCS domain. The four metal binding residues are highlighted along with other residues which were mutated for the transcription assay. D. Overlay of hPh1 FCS (red), L3MBTL2 FCS (yellow, PDBID: 2W0T) (16), and the bacterial YacG peptide (cyan, PDBID: 1LV3) (30). The overlay shows the greatest variability of this fold occurs in the loop region.