Table 1. Optimized parameters of the mathematical Kdp model.
| Parameter name | Value | Units | Description | ||
| Wild type | E. coli RH010 | E. coli RH010/pKT84 | |||
| Two-component system | |||||
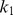
|
0.23 |

|
Autophosphorylation of D, forward reaction rate constant | ||
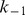
|
5.1×10−6 |
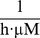
|
Autophosphorylation of D, backward reaction rate constant | ||
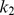
|
2.27×103 |

|
Phosphotransfer to E, forward reaction rate constant | ||
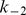
|
8.7×10−4 |
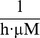
|
Phosphotransfer to E, backward reaction rate constant | ||

|
40.6×10−3 |
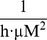
|
Dephosphorylation of EP by D | ||
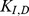
|
520 | mM | Inhibition of autophosphorylation of D by free K+ | ||
| Transcription | |||||
| K | 4×104 | 1.2×104 | 6×104 | 1 | Equilibrium binding constant of σ-factor and RNA polymerase to DNA |
| KE | 5.32×10−2 |
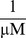
|
DNA-binding of free EP, equilibrium dissociation constant | ||
| α | 2.59×10−3 |

|
Affinity factor | ||
| ktr | 1.06×104 |

|
Transcription rate constant | ||
| kz | 21.74 |

|
Transcript degradation rate constant | ||
| Translation | |||||
| ktl1 | 5.4 |

|
Translation rate constant of D | ||
| ktl2 | 162 |

|
Translation rate constant of E | ||
| ktl3 | 8.1×103 |

|
Translation rate constant of F | ||
| kd,F | 4.8 | 11.4 | 4.8 |

|
Degradation rate constant of F |
| kd | 0.2 |

|
Degradation rate constant of D and E | ||
| Potassium pools | |||||
| kKdp | 7.86×103 | 0.46×103 |

|
K+ uptake rate constant; given that  at steady state and cell dry weight is DW = 2.8×10−13 g, one obtains an estimate of at steady state and cell dry weight is DW = 2.8×10−13 g, one obtains an estimate of 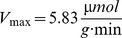 ; in the literature we find ; in the literature we find 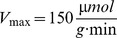 [56]
[56]
|
|
| KM,Kdp | 3.83 | 4 | mM | Half saturation constant of K+ uptake; literature value for Kdp: KM = 2 µM | |
| KI,Kdp | 100 | 0 | mM | Inhibition of K+ uptake by free K+ | |
| Vmax,Trk | 36.5 |
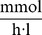
|
Maximum velocity of K+ uptake by Trk | ||
| KM,Trk | 0.1 | mM | Half saturation constant of K+ uptake by Trk | ||
| Vmax,Ktr | 0 | 400 |
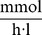
|
Maximum velocity of K+ uptake by KtrAB | |
| KM,Ktr | 0 | 5×10−2 | mM | Half saturation constant of K+ uptake by KtrAB; | |
| Vmax,lys | 1×10−2 | 2×10−2 | 1×10−2 |
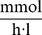
|
Maximum K+ release rate due to cell lysis |
| KM,lys | 150 | mM | Half saturation constant of K+ release due to cell lysis | ||
| kbind | 8 |

|
Binding rate constant of free K+ | ||
| KM,free | 250 | mM | |||
| kdiss | 7.81 |

|
Dissociation rate constant of bound K+ | ||
| τ | 0 | 0.35 | 0 | h | “Delay” constant for intracellular K+ exchange |
| Growth | |||||
| kμ,1 | 0.54 | 0.59 | 0.54 |

|
Maximum growth rate |
| kμ,2 | 1.43×10−3 | 1.52×10−3 | 1.57×10−3 |

|
Carrying capacity, inflection point of growth curve |

|
6 | 1 | 6 | 1 | Determines maximum steepness of growth curve |
The experimental data for the wild type, the RH010 mutant and the RH010/pKT84 mutant cannot be reproduced using a single set of parameters. The Table lists the values of each parameter used to describe the dynamics of each strain.
