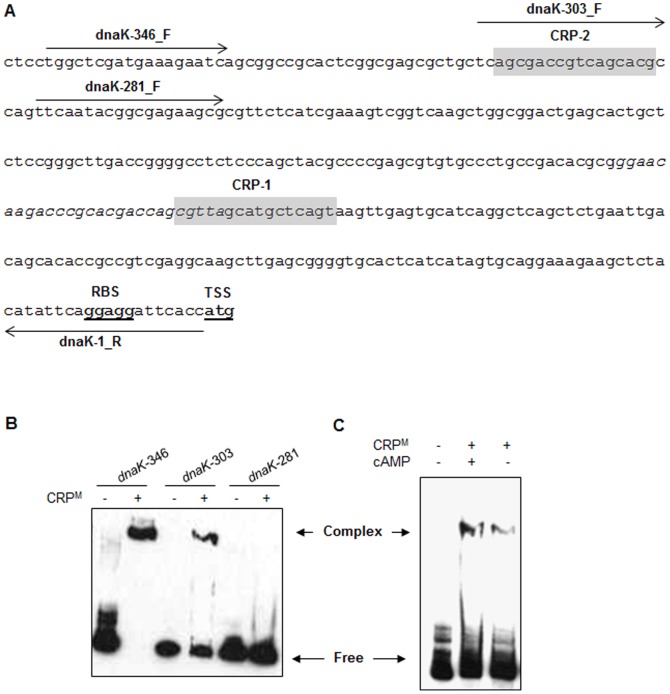
Figure 4. Identification of CRP-binding motif in promoter region of dnaK operon by EMSA.
A) Sequence of the 5'-UTR of dnaK operon. The putative ribosome-binding site (RBS), and the translation start site (TSS) are boldfaced and underlined. The putative CRP-binding sites are shaded, whereas the SigH-recognition sequence is shown in italics. Positions and directions of the primers that were used for PCR amplification of DNA fragments shown in (B) are marked by horizontal arrows. B) Interaction of CRPM with the 5'-UTR of dnaK operon. Biotin-labeled dnaK promoter fragments of various lengths, as shown, were incubated with CRPM in the presence of 1 mM cAMP before separation of DNA-protein complexes by gel electrophoresis. Reaction mixtures containing promoter fragments but lacking the CRPM were resolved in adjacent lanes as controls. C) Effect of cAMP on complex formation between CRPM and dnaK promoter. Purified CRPM pre-incubated with or without 1 mM cAMP for 30 min on ice, was subject to binding with full length dnaK promoter fragment (dnaK-346) for 15 min at 37°C before separation of CRPM–dnaK complexes by gel electrophoresis. Complex, CRPM–dnaK complex; Free, unbound DNA fragment.