Table 1. The rare variants identified in DLC1 isoform 1.
| Variant type | Patient ID | Gender | Age of diagnosis | Diagnosis | Exon | Nucleotide alterationa | Amino acid alteration | SIFT score | SIFT prediction (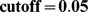 ) ) |
Number of mutations in patients | Number of mutations in controls | In dbSNPb | ALT allele frequency in dbSNPc |
| Private variants | 67 | M | 5 | VSD&PFO | 2 | c.797G>A | p.Gly266Glu | 0.406 | Tolerated | 1/151 | 0/900 | Na | Na |
| 153 | F | 8 | VSD | 3 | c.1048G>A* | p.Ala350Thr | 0.368 | Tolerated | 1/151 | 0/900 | Na | Na | |
| 168 | F | 5 | ASD | 3 | c.1079T>A* | p.Met360Lys | 0.001 | Damaging | 1/151 | 0/900 | Na | Na | |
| 169 | F | 22 | PS | 4 | c.1252G>A* | p.Glu418Lys | 0.027 | Damaging | 1/151 | 0/900 | Na | Na | |
| 89 | F | 2 | PDA | 4 | c.1298C>A | p.Thr433Asn | 0.02 | Damaging | 1/151 | 0/900 | Na | Na | |
| 131 | F | 8 | PDA | 9 | c.[1661A>T(+)1662T>C]* | p.Asp554Val | 0.014 | Damaging | 1/151 | 0/900 | Na | Na | |
| 190 | F | 7 | VSD | 9 | c.[1661A>T(+)1662T>C]* | p.Asp554Val | 0.014 | Damaging | 1/151 | 0/900 | Na | Na | |
| Other rare variants | 49 | F | 9 | TOF | 2 | c.659C>T | p.Ala220Val | 1 | Tolerated | 1/151 | 1/900 | Na | Na |
| 61 | F | 6 | TOF | 3 | c.1051C>T | p.Arg351Trp | 0 | Damaging | 1/151 | 2/900 | rs144283917 | 2.324/5869 | |
| 42 | F | 17 | VSD | 4 | c.1237T>A* | p.Leu413Met | 0.005 | Damaging | 1/151 | 0/900 | rs143447199 | 1/4545 | |
| 55 | F | 26 | PDA | 9 | c.1683C>A | p.Asp561Glu | 0.171 | Tolerated | 1/151 | 2/900 | rs201661577 | 5/2174 | |
| 124 | F | 4 | VSD | 9 | c.2854C>G* | p.Leu952Val | 0.003 | Damaging | 1/151 | 0/900 | rs184157214 | 1/2000 | |
| 28 | M | 1 | VSD | 16 | c.4111G>C* | p.Val1371Leu | 0.016 | Damaging | 1/151 | 0/900 | rs142865083 | 1/2000 | |
| 8 | M | 12 | VSD | 18 | c.4533C>G | p.Ile1511Met | 0.001 | Damaging | 1/151 | 0/900 | rs78322853 | Na |
Note. Na, no available data; M, male; F, female; VSD, ventricular septal defect; PFO, patent foramen ovale; ASD, atrial septal defect; PS, pulmonary stenosis; PDA, patent ductus arteriosus; TOF, tetralogy of Fallot. a, Nucleotide numbering is according to the RefSeq database NM_182643.2. b, The version of dbSNP used in the table is dbSNP build 137. c, The alternative allele frequency form the dbSNP database is calculated by the alternative allele count/two times the number of individuals assayed. *The mutant vectors were constructed according to these variants.
