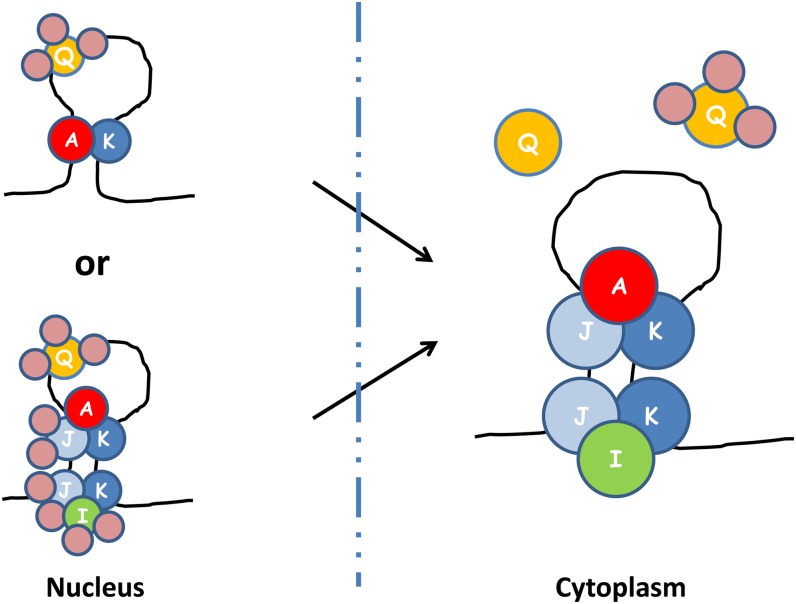
Figure 8.
A proposed model of prolamine zipcode mRNA-RBP protein assembly complexes based on results obtained by IP, yeast two-hybrid analysis, and BiFC. Note that at least three different RBP complexes bind to the prolamine zipcodes to form two ribonucleoprotein assembly complexes. RBP-A, RBP-J, and RBP-K and RBP-I, RBP-J, and RBP-K are assembled in the cytoplasm as two separate multiprotein complexes on the prolamine zipcode. When associated with RBP-A and RBP-K, RBP-Q is not accessible to its antibody, suggesting that it is bound by other proteins that comprise a third multiprotein family. As BiFC results indicate that heterodimer formation occurs in the nucleus (Figure 7), it is likely that multiprotein complexes A-J-K and I-J-K are formed in the nucleus, where they interact with the prolamine zipcode. If so, RBP-I and RBP-J are bound by other proteins so that they are not accessible to their respective antibodies. A simpler ribonucleoprotein assembly complex in the nucleus would consist of two multiprotein complexes, one containing RBP-Q and a second containing RBP-A and RBP-K. Note that the various multiprotein complexes were arbitrarily drawn on the zipcode stem-loop structure. The actual binding specificity of the various multiprotein complexes to the stem-loop structure remains to be resolved.