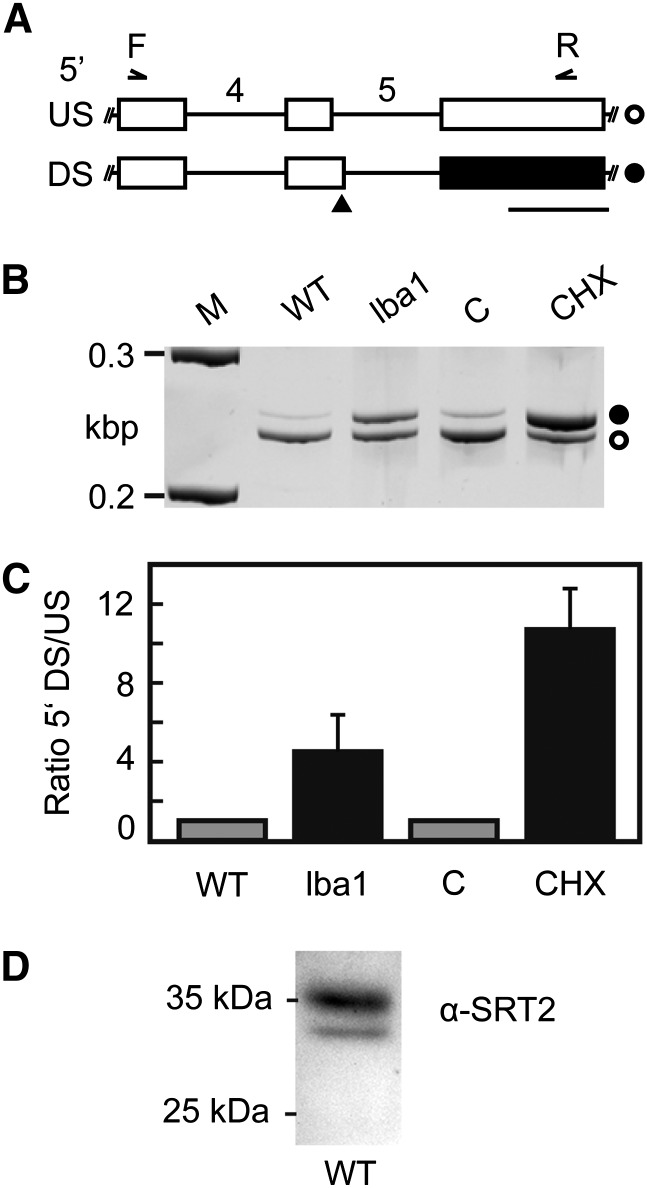
Figure 1.
AS of SRT2 precursor mRNA generates an NMD target and is regulated by polypyrimidine tract-binding protein splicing factors. A, Partial models of SRT2 splicing variants resulting from usage of an upstream (US, top) or downstream (DS, bottom) 5′ splice site in intron 5 (introns numbered). Boxes and lines depict exons and introns, respectively. White boxes correspond to coding sequence, and black box indicates an untranslated region resulting from the introduction of a premature termination codon (black triangle) upon usage of the downstream 5′ splice site. White circle indicates SRT2 splice form 1,2,3, and 7, and black circle indicates splice form 4 and 6. Binding positions of primers used for coamplification of splicing variants in B and C are shown. Bar = length of 100 bp. B, RT-PCR analysis of AS event shown in A. For NMD impairment, low-beta-amylase1 (lba1) mutant or cycloheximide (CHX) versus mock (C) treatment were analyzed. M indicates ladder with 0.2- and 0.3-kb DNA fragments. C, Quantitative analysis of splicing ratios for samples depicted in B using a bioanalyzer. Displayed are mean values (n = 3–6, ± sd). Data are normalized to the wild type (WT) or mock treatment. D, Western-blot analysis of SRT2 protein in Arabidopsis leaves.