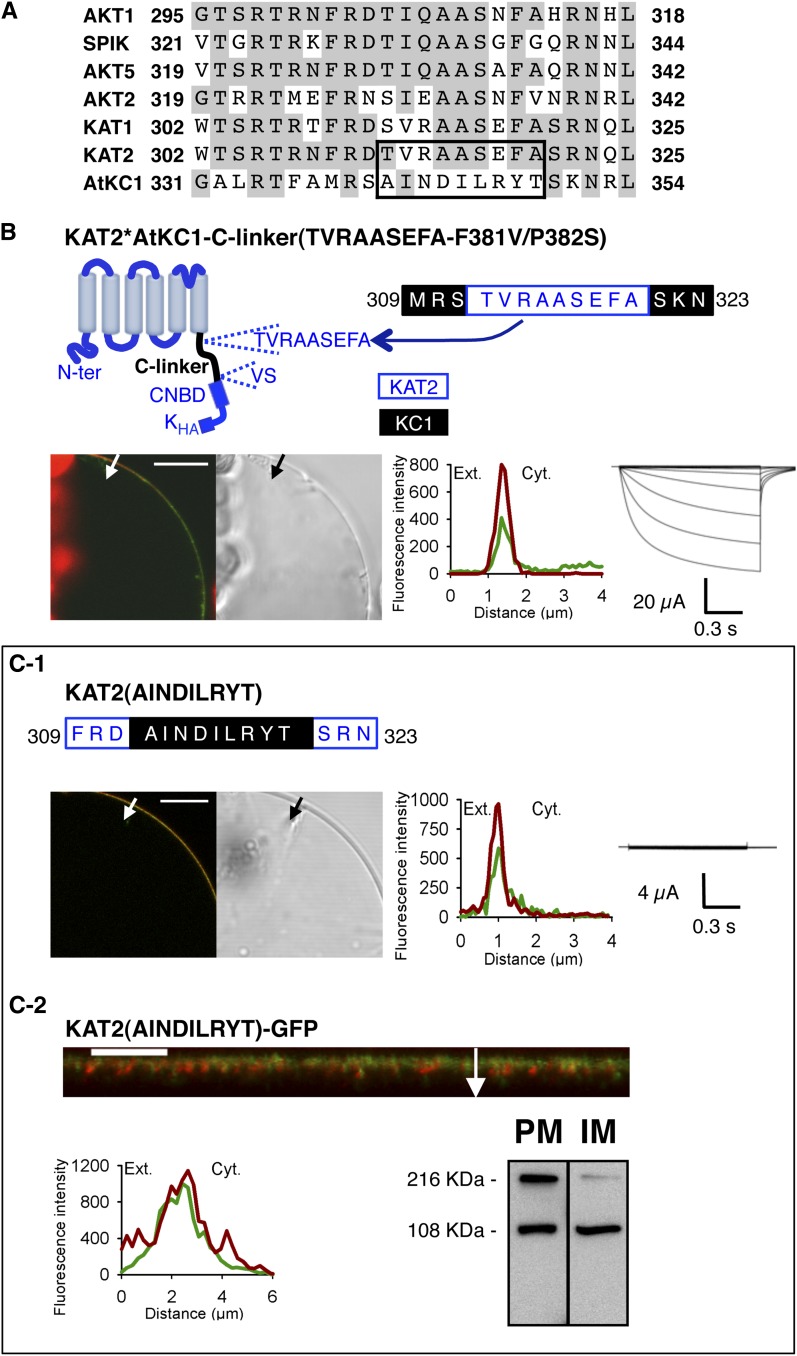
Figure 6.
The TVRAASEFA stretch present in the N-terminal region of the KAT2 C-linker plays an essential role in KAT2 channel functionality. A, Alignment displaying the first 24 amino acids of the C-linker from all Arabidopsis inward Shaker channel subunits. Identical residues in at least three members are displayed in gray background. The TVRAASEFA stretch of KAT2 (amino acids 312–320) and its counterpart in AtKC1 (amino acids 341–349) are boxed. B, The KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) chimera is active at the PM. Top left, pictogram of this chimera showing the sequences from KAT2 (in blue) and AtKC1 (in black). This chimera was obtained from the KAT2*AtKC1-C-linker(F381V/P382S) one (Fig. 3H) by replacing the AINDILRYT stretch from the AtKC1 C-linker by the KAT2 TVRAASEFA sequence. The position of this substitution in the C-linker sequence of the resulting KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) chimera is indicated at the top right (KAT2 amino acids in blue and AKC1 amino acids in black). C, Substituting the AtKC1 sequence AINDILRYT for the TVRAASEFA motif in KAT2 renders the channel inactive. The position of this substitution in the C-linker sequence of the resulting mutant, named KAT2(AINDILRYT), is indicated at the top in C1 (KAT2 amino acids in blue and AKC1 amino acids in black). In B and C1, surface expression and activity at the PM of the two constructs were investigated by subcellular localization of GFP fusions in tobacco protoplasts and current recordings in X. laevis oocytes, as described in the legend to Figure 1. In C2, further analyses were performed in oocytes expressing KAT2(AINDILRYT)-GFP to check its subcellular localization by confocal imaging and protein gel-blot analysis of cellular fractions, as described in the legend to Figure 1. A white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm. Bars = 10 μm.