Cytokinin protects plants from the consequences of light stress by acting on reactive oxygen species scavenging and D1 protein levels, thereby preventing photoinhibition.
Abstract
Cytokinins are plant hormones that regulate diverse processes in plant development and responses to biotic and abiotic stresses. In this study, we show that Arabidopsis (Arabidopsis thaliana) plants with a reduced cytokinin status (i.e. cytokinin receptor mutants and transgenic cytokinin-deficient plants) are more susceptible to light stress compared with wild-type plants. This was reflected by a stronger photoinhibition after 24 h of high light (approximately 1,000 µmol m−2 s−1), as shown by the decline in maximum quantum efficiency of photosystem II photochemistry. Photosystem II, especially the D1 protein, is highly sensitive to the detrimental impact of light. Therefore, photoinhibition is always observed when the rate of photodamage exceeds the rate of D1 repair. We demonstrate that in plants with a reduced cytokinin status, the D1 protein level was strongly decreased upon light stress. Inhibition of the D1 repair cycle by lincomycin treatment indicated that these plants experience stronger photodamage. The efficiency of photoprotective mechanisms, such as nonenzymatic and enzymatic scavenging systems, was decreased in plants with a reduced cytokinin status, which could be a cause for the increased photodamage and subsequent D1 degradation. Additionally, slow and incomplete recovery in these plants after light stress indicated insufficient D1 repair. Mutant analysis revealed that the protective function of cytokinin during light stress depends on the ARABIDOPSIS HISTIDINE KINASE2 (AHK2) and AHK3 receptors and the type B ARABIDOPSIS RESPONSE REGULATOR1 (ARR1) and ARR12. We conclude that proper cytokinin signaling and regulation of specific target genes are necessary to protect leaves efficiently from light stress.
Light absorption, the subsequent conversion into biochemical energy, and the production of oxygen of plants play an essential role for life on earth. Although light is a prerequisite for this process, high light (HL) easily exceeds the plant’s capacity to assimilate CO2, causing an overreduction of the electron transport chain that results in the inactivation of PSII (photoinhibition; Barber and Andersson, 1992; Aro et al., 1993; Yamamoto et al., 2008). One of the major targets of photoinhibition is the PSII core protein D1 (for review, see Adir et al., 2003; Edelman and Mattoo, 2008).
Damaged D1 proteins are continuously replaced by de novo-synthesized D1 in a process called the D1 repair cycle (Aro et al., 1993, 2005; Baena-González and Aro, 2002). This cycle consists of (1) migration of damaged D1 protein from the grana to the stroma lamellae, (2) proteolytic degradation of damaged D1 protein by FILAMENTATION TEMPERATURE SENSITIVE H (FTSH) protease and DEGRADATION OF PERIPLASMIC PROTEINS PROTEASE (DEGP), (3) de novo synthesis of precursor D1 protein (preD1) and cotranslational insertion into the thylakoid membrane, (4) C-terminal processing of preD1 catalyzed by the C-TERMINAL PEPTIDASE (CTP), and (5) migration to the grana thylakoids and formation of a fully functional PSII complex (for review, see Kato and Sakamoto, 2009). The D1 repair cycle enables a high D1 turnover, which is indispensable, especially under HL conditions. Photoinhibition always occurs when the rate of damage exceeds the D1 repair capacity. Besides causing photoinhibition, excess light also provokes the generation of reactive oxygen species (ROS) as by-products of photosynthesis (Asada, 1999; Apel and Hirt, 2004). In PSI, the reduction of oxygen leads to the formation of superoxide anion radicals that can be converted into hydrogen peroxide (H2O2) and hydroxyl radicals, whereas in PSII, the transfer of excitation energy from excited triplet chlorophyll (3chl*) to oxygen results in the production of singlet oxygen (1O2; Fischer et al., 2013). ROS were considered to accelerate photoinhibition by causing direct photooxidative damage of the D1 protein. However, it was shown that acceleration of photoinhibition by ROS actually results from an inhibition of the D1 repair rather than from the direct damage of D1 (for review, see Takahashi and Murata, 2008). In order to reduce ROS levels, plants have developed an efficient scavenging system composed of enzymatic and nonenzymatic scavengers. Enzymatic scavengers such as superoxide dismutase (SOD), ascorbate peroxidase (APX), and catalase detoxify superoxide anion radicals and H2O2. The nonenzymatic antioxidants such as glutathione, ascorbate, tocopherol, and carotenoids including the xanthophylls are important for the elimination of hydroxyl radicals and 1O2 and the quenching of lipid peroxy radicals and 3chl* (Havaux et al., 2005). All these scavenging systems are important protective mechanisms of plants against photoinhibition and minimize the inhibition of D1 repair (Nishiyama et al., 2001; Takahashi et al., 2007).
In addition to an efficient ROS-scavenging system, plants have developed other protective mechanisms to avoid photoinhibition (for review, see Takahashi and Badger, 2011), such as the dissipation of excess light energy as thermal energy, cyclic electron transport, and light-avoidance movements of chloroplasts and leaves. These processes minimize the risk of the overreduction of photosynthetic antenna complexes and core proteins as well as the inhibition of D1 repair and hence photoinhibition of PSII.
Cytokinins are a class of plant hormones that regulate numerous developmental processes such as the cell cycle, shoot and root growth, seed germination, and leaf senescence (for review, see Werner and Schmülling, 2009). In Arabidopsis (Arabidopsis thaliana), cytokinins are perceived by three membrane-located receptors, the His kinases ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and CYTOKININ RESPONSE1 (CRE1)/AHK4 (Inoue et al., 2001; Suzuki et al., 2001), which have distinct functions (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006; for review, see Heyl et al., 2012). The signal is further transmitted by a two-component signaling system via HISTIDINE PHOSPHOTRANSFER PROTEINS (named AHPs in Arabidopsis) to type B RESPONSE REGULATORS (type B ARRs in Arabidopsis), which are transcription factors and regulate cytokinin response genes (Heyl and Schmülling, 2003; Müller and Sheen, 2007).
Cytokinin influences several light-regulated processes. They can partially mimic photomorphogenesis in etiolated seedlings (Chory et al., 1994; Lochmanová et al., 2008), which is mediated by the transcription factors ARR1, ARR10, and ARR12 (Argyros et al., 2008). Cytokinin acts as a signal for photosynthetic acclimation to canopy light gradients (Boonman et al., 2007) and shade-induced leaf growth arrest (Carabelli et al., 2007). Exogenous application of cytokinin stimulates the transition from etioplast to chloroplast in detached leaves and cell cultures, increases the rate of grana and stroma lamella, and extends the life span of chloroplasts (Parthier, 1979; Catský et al., 1993; Mok, 1994; Cherniad’ev, 2000; Synková et al., 2006). Furthermore, cytokinin influences photosynthesis and related processes (Kusnetsov et al., 1998; Synková et al., 1999; Yaronskaya et al., 2006; Cortleven and Valcke, 2012), which is at least partially due to the control of gene expression (Rashotte et al., 2003; Brenner et al., 2005; Zubo et al., 2005, 2008).
Recent research has provided evidence that cytokinins are also involved in stress responses (for review, see Argueso et al., 2009; Ha et al., 2012). For instance, cytokinins are known to induce an antioxidant protection mechanism in chloroplasts (Procházková et al., 2008) and alter the transcript levels of many stress-inducible genes (Rashotte et al., 2003; Brenner et al., 2005, 2012; Brenner and Schmülling, 2012; Bhargava et al., 2013). Several reports revealed a role for cytokinins during drought, cold, and osmotic stress (Rivero et al., 2007; Tran et al., 2007; Jeon et al., 2010; Nishiyama et al., 2011; Macková et al., 2013). However, a role for cytokinin during light stress has not yet been described.
The availability of Arabidopsis plants with a reduced cytokinin status (i.e. plants with a lower cytokinin content or signaling) strongly improves the possibility to explore novel functions of cytokinin. The knockout of cytokinin receptors is one option to obtain plants with a reduced cytokinin status, and mutants lacking one, two, or all three receptors have been described (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Alternatively, a reduced cytokinin content can be generated by the overexpression of CKX genes, encoding cytokinin oxidases/dehydrogenases catalyzing cytokinin degradation (Werner et al., 2001, 2003). In this study, we used both the cytokinin receptor double mutant ahk2 ahk3 and cytokinin-deficient 35S:CKX4 transgenic plants to study the consequences of a reduced cytokinin status for the response to light stress in leaves. The data revealed that cytokinin is an essential factor for protection against light stress, since plants with a reduced cytokinin status are more sensitive to excess light. They exhibited a decreased photosynthetic activity in comparison with the wild type which was accompanied by increased photodamage after exposure to 24 h of HL. These results indicate that several photoprotective mechanisms were impaired in cytokinin-deficient plants, causing a strong increase in photodamage exceeding the efficiency of the D1 repair and leading to photoinhibition.
RESULTS
A Reduced Cytokinin Status Causes Increased Photooxidative Stress and Reduced Photosynthetic Activity after HL Treatment
We repeatedly observed that mutants and transgenic Arabidopsis plants with a reduced cytokinin status showed higher sensitivity to light than wild-type plants. The cytokinin receptor double mutant ahk2 ahk3 and 35S:CKX transgenic plants (here we used 35S:CKX4-overexpressing plants) appeared to be particularly sensitive and, therefore, were exposed to defined light stress conditions. Plants were grown for 28 to 32 d under short-day (SD) conditions and then exposed to HL (1,000 µmol m−2 s−1) for 24 h.
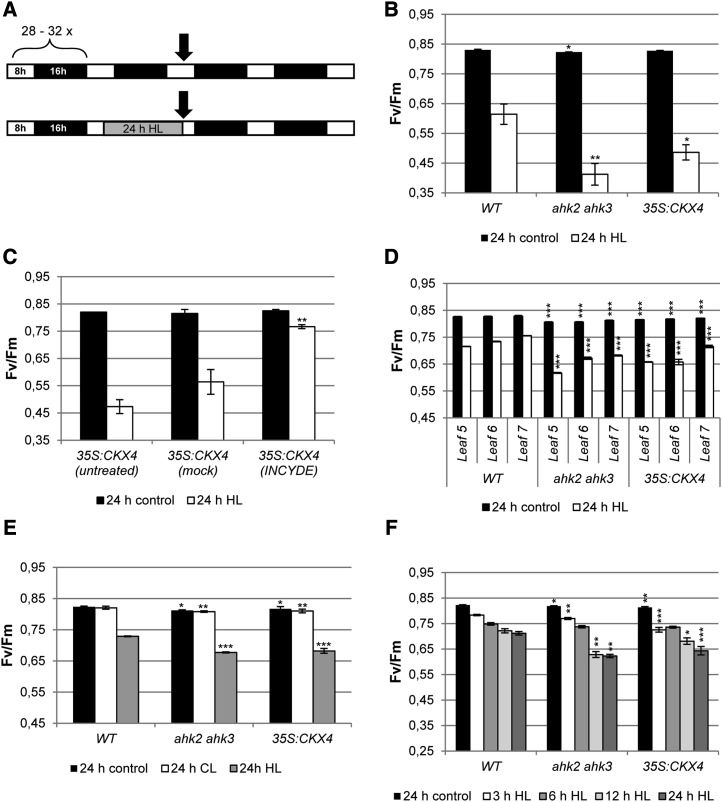
The ratio of variable fluorescence to maximum fluorescence (Fv/Fm) was analyzed immediately after the HL treatment, as indicated in Figure 1A. A strong decrease in Fv/Fm was measured in cytokinin-deficient plants (35S:CKX4 and ahk2 ahk3), while wild-type plants showed a much weaker response (Fig. 1B). Treatment with the CKX inhibitor INCYDE (Zatloukal et al., 2008) prior to stress application dramatically reduced photoinhibition in cytokinin-deficient 35S:CKX4 plants, as shown by only a minor decrease in Fv/Fm in comparison with mock-treated plants (Fig. 1C).
Figure 1.
HL treatment reduces the photochemical efficiency of PSII in plants with a reduced cytokinin status. A, Schematic overview of the experimental design. Soil-grown 4-week-old plants were continuously grown under SD conditions or exposed for 24 h to HL. White bars, Light period; black bars, dark period; gray bar, HL period. Arrows indicate sampling points. B, Photochemical efficiency of PSII (Fv/Fm) in whole plants (leaf 6) after 24 h of HL (n = 8). C, Fv/Fm in whole 35S:CKX4 plants (leaf 6) sprayed once per day with 10 µm INCYDE starting 5 d before HL treatment (n = 6). D, Fv/Fm in detached leaves 5, 6, and 7 (n = 8). E, Fv/Fm in detached leaf 7 directly after 24 h of light treatment (continuous moderate light [CL] or HL; n = 8). F, Fv/Fm of detached leaf 6 after different HL exposure times (n = 8). Asterisks indicate significant differences from the respective wild-type (WT) condition (* 0.05 > P > 0.01; ** 0.01 > P > 0.001; *** P < 0.001). Error bars represent se.
In order to investigate the role of cytokinin during HL stress in more detail, further experiments were carried out with detached leaves. Similar to the experiments performed on whole plants, HL treatment triggered a stronger reduction of Fv/Fm in cytokinin-deficient leaves compared with wild-type plants. This response was independent of the leaf age (Fig. 1D). Importantly, the stress response was due to the HL treatment and not caused by the lack of the dark period, as continuous moderate light (instead of HL) did not result in reduction of Fv/Fm values (Fig. 1E). Kinetics of the HL stress response (measured as Fv/Fm) revealed that the divergence between cytokinin-deficient and wild-type plants already started after 3 h of HL treatment, but the difference was more pronounced after 12 and 24 h of illumination (Fig. 1F).
The D1 Protein Level Is Strongly Reduced by Light Stress in Plants with a Reduced Cytokinin Status
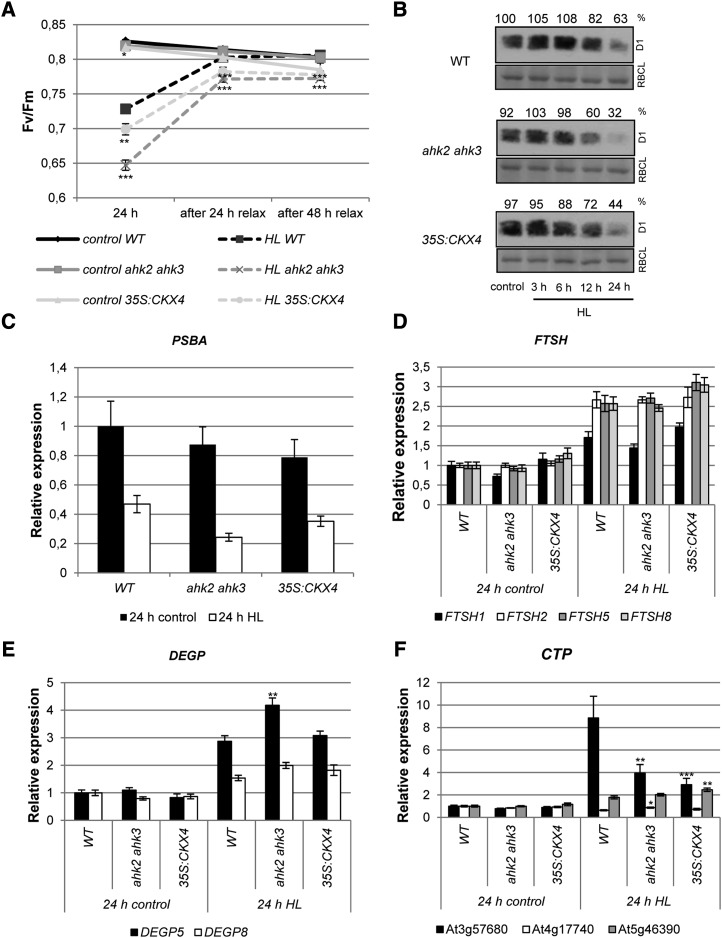
The lower photochemical efficiency of cytokinin-deficient plants after exposure to HL could be either the consequence of a hampered D1 repair cycle or a higher degree of photodamage (i.e. damaged PSII proteins) or both. During relaxation under SD conditions, photosystems are able to recover from light stress, which can be monitored by the increase in Fv/Fm. In ahk2 ahk3 and 35S:CKX4 plants, the recovery capacity was attenuated compared with the wild type, as indicated by significantly lower Fv/Fm values after 24 and 48 h of relaxation (Fig. 2A). Fv/Fm did not return to initial levels in these plants, which had been the case for wild-type plants after 48 h of relaxation (Fig. 2A).
Figure 2.
Effects of HL treatment on the functionality and protein and transcript levels of genes encoding D1 and proteases of the D1 repair cycle. Defined detached leaves of 4-week-old Arabidopsis plants were exposed for 24 h to continuous HL. A, Fv/Fm of leaf 6 after 24 h of HL and relaxation for 24 and 48 h (n = 22). B, Analysis of the D1 protein level. Protein was extracted from pooled leaves 5, 6, and 7 after different HL exposure times. Numbers above the blots indicate D1 protein level as a percentage of the wild-type (WT) control (n = 3). Representative blots are shown. RBCL, Large subunit of Rubisco. C to F, Analysis of transcript levels by quantitative real-time PCR of the PSBA (C), FTSH (D), DEGP (E), and CTP (F) genes. Transcript levels of wild-type leaves under control conditions were set to 1 (n = 8). Asterisks indicate significant differences from the respective wild-type condition (* 0.05 > P > 0.01; ** 0.01 > P > 0.001; *** P < 0.001). Error bars represent se.
The D1 protein is the most vulnerable component of PSII, and its replacement by newly synthesized functional D1 protein is required to avoid photoinhibition. To explore whether the D1 protein level was differently affected in wild-type plants and in plants with a reduced cytokinin status, we determined the D1 protein abundance. Additionally, transcript levels of the PHOTOSYSTEM II REACTION CENTER PROTEIN A (PSBA) gene encoding the D1 protein and various genes encoding proteins of the D1 repair cycle were analyzed. The D1 protein level was investigated by protein gel-blot analysis (Fig. 2B). D1 protein levels were almost unchanged in all genotypes after 3 to 6 h of HL treatment. However, after longer HL treatment, the D1 protein level was strongly diminished in leaves of ahk2 ahk3 and 35S:CKX4 plants in comparison with leaves of wild-type plants. The strongest difference in D1 protein levels between the cytokinin-deficient and wild-type plants was noted after 24 h of HL. In the wild type, 63% of the initial D1 protein level was retained, while only 32% and 44% of the D1 protein level compared with the wild-type control were left in ahk2 ahk3 and 35S:CKX4, respectively. This is consistent with the kinetics of Fv/Fm reduction shown in Figure 1F, revealing the largest differences after 12 and 24 h of HL treatment. Although no statistically significant differences were observed, a clear trend was visible: cytokinin-deficient plants showed a stronger decrease in PSBA transcript level after 24 h of HL in comparison with the wild type (Fig. 2C).
Next, we analyzed whether the expression of genes involved in the D1 repair cycle, such as FTSH (FTSH1, FTSH2, FTSH5, and FTSH8; Fig. 2D), DEGP (DEGP5 and DEGP8; Fig. 2E), and CTP homologs (At4g17740, At5g46390, and At3g57680; Fig. 2F), was altered in plants with a lowered cytokinin status. FTSH and DEGP encode proteases involved in D1 degradation (Kato and Sakamoto, 2009). Most of these transcripts were induced upon HL treatment, but generally, the basic steady-state mRNA levels and the degree of induction were similar in cytokinin-deficient and wild-type plants. One exception is DEGP5, which was more strongly induced in ahk2 ahk3 mutants in comparison with the wild type and 35S:CKX4 (Fig. 3E). Three Arabidopsis genes (At3g57680, At4g17740, and At5g46390) have been predicted to encode CTP homologs (Satoh and Yamamoto, 2007; Yin et al., 2008), based on the similarity of their amino acid sequences with the C-terminal processing peptidase encoded by the cyanobacterial CtpA gene and required for maturation of the D1 protein (Fig. 2F). Among these, At3g57680 showed a strong (9-fold) induction in leaves of wild-type plants in response to light stress. At5g46390 showed a slight induction (2-fold), while the third homolog did not respond to HL treatment (Fig. 2F). The transcriptional response of the latter two genes was similar in cytokinin-deficient plants to that in the wild type. However, in contrast to the strong up-regulation of At3g57680 in the wild type, cytokinin-deficient plants showed only a 3- to 4-fold induction (Fig. 2F). The HL response of the corresponding ctpa1 mutant (Yin et al., 2008) was analyzed. This mutant behaved like the wild type (data not shown), which indicates that At3g57680 has no important function in the light stress response and that its different regulation is probably not the reason for the HL phenotype in cytokinin-deficient plants.
Figure 3.
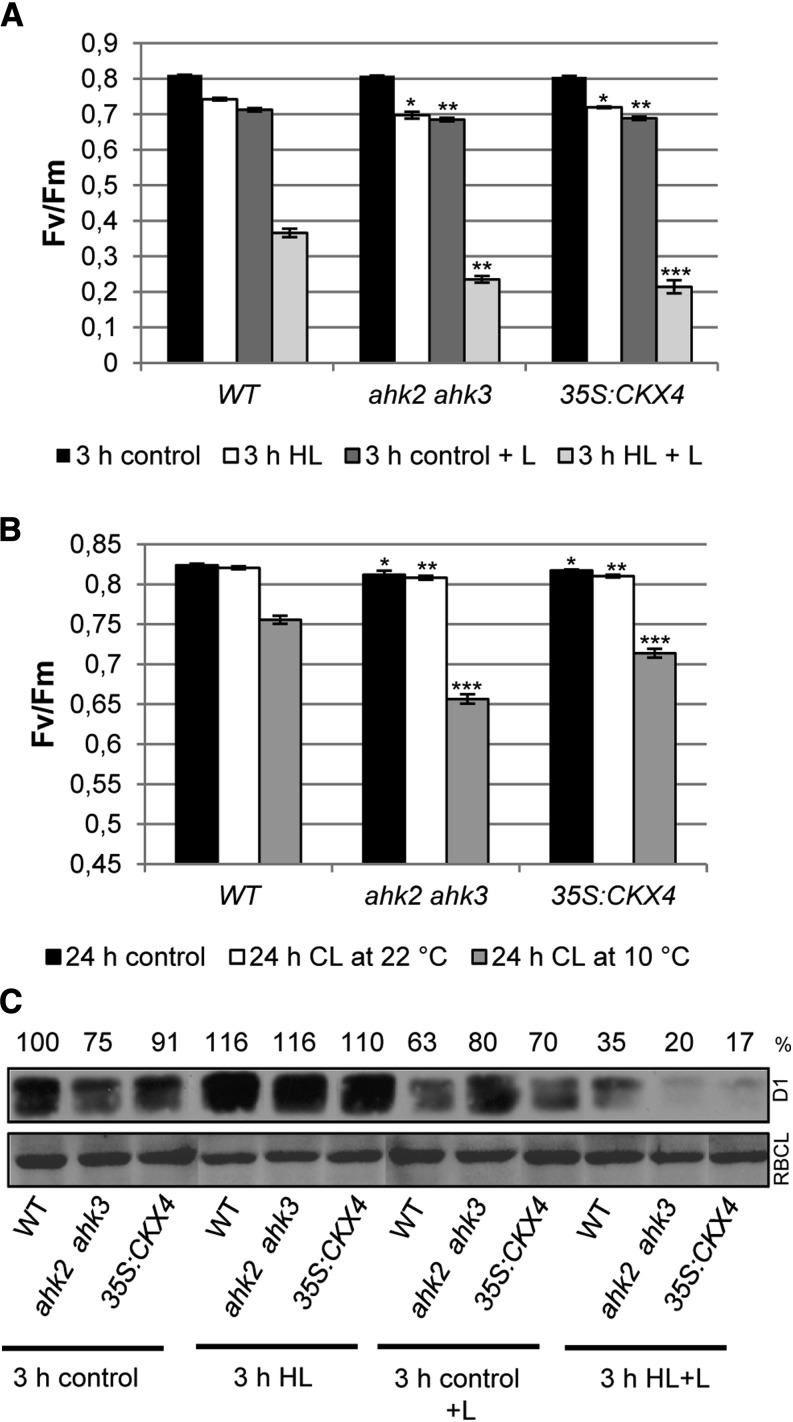
Light stress increases photodamage in plants with a reduced cytokinin status. Photochemical efficiency was measured in detached leaves of 4-week-old Arabidopsis plants. A, Fv/Fm of leaf 6 irradiated for 3 h with HL in the absence or presence of 1 mm lincomycin (+ L). B, Fv/Fm of leaf 7 treated for 24 h with continuous light (CL) at 22°C and 10°C. Asterisks indicate significant differences from the respective wild-type condition (* 0.05 > P > 0.01; ** 0.01 > P > 0.001; *** P < 0.001). Error bars represent se (n = 8). C, Blot analysis of D1 protein in leaf material derived from the experiment shown in A. Numbers above the blots indicate D1 protein level as a percentage of the wild-type (WT) control (n = 4). Representative blots are shown. RBCL, Large subunit of Rubisco.
Photodamage Is Increased in Plants with a Reduced Cytokinin Status
To evaluate whether an increased photodamage contributes to the light stress phenotype of cytokinin-deficient plants, we inhibited the D1 repair cycle by two different means. Detached leaves were treated with lincomycin, which is an inhibitor of plastid protein synthesis and thus completely blocks the D1 repair cycle, or they were exposed to low temperature, which also reduces D1 repair activity (Grennan and Ort, 2007; Mohanty et al., 2007). This inhibition enables one to exclusively monitor the damage of PSII. Both treatments caused a strong decrease of Fv/Fm in all genotypes, demonstrating the importance of the D1 repair cycle for protecting against HL stress (Fig. 3, A and B). The stronger reduction of Fv/Fm in ahk2 ahk3 and 35S:CKX4 as compared with the wild type indicated that cytokinin-deficient plants experienced a higher degree of photodamage, which further demonstrates that they encounter increased photooxidative stress. This higher degree of photodamage in cytokinin-deficient plants is also reflected by a more severe decline in D1 protein, which decreased to almost only half of the level in the wild type after HL plus lincomycin treatment (Fig. 3C, 3 h HL+L).
In conclusion, an elevated level of photodamage in cytokinin-deficient plants appears to be an important cause for the more pronounced HL response and the succeeding lack of recovery in these plants.
Plants with a Reduced Cytokinin Status Show Reduced ROS-Scavenging Capacity
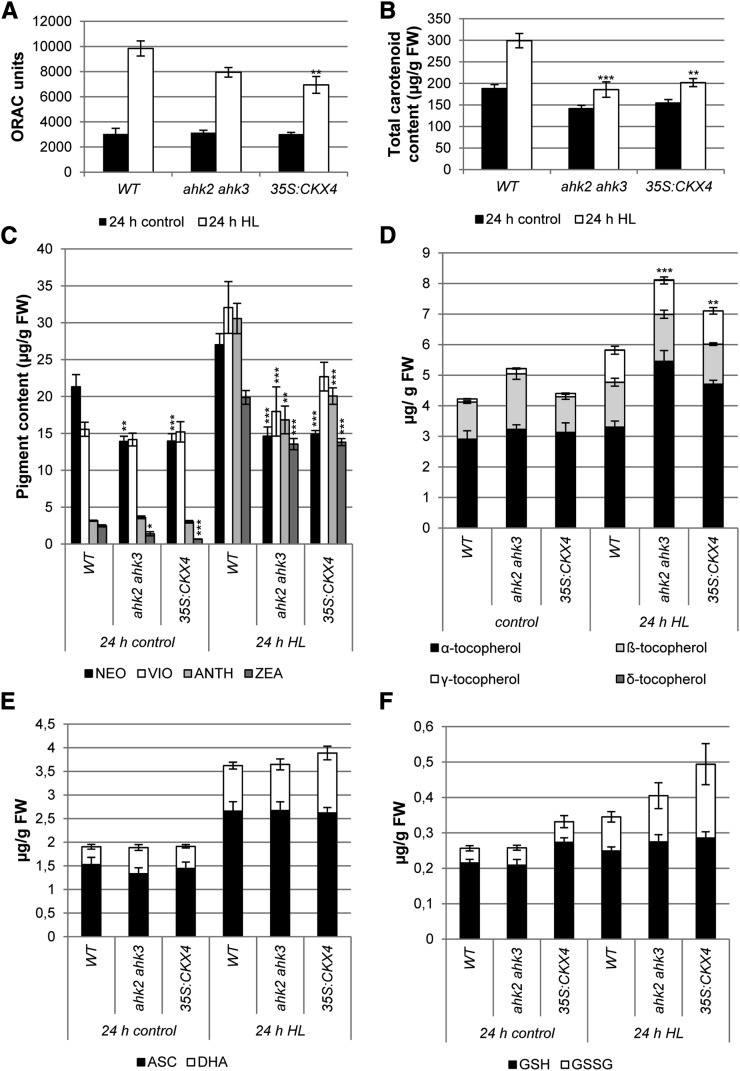
The higher degree of photodamage in plants with a reduced cytokinin status could be due to a reduced efficiency of scavenging mechanisms. To further test this hypothesis, the total antioxidant capacity was determined by the oxygen radical antioxidant capacity (ORAC) assay. This initial experiment indicated that the ahk2 ahk3 and 35S:CKX4 plants had about 20% to 30% lower total antioxidant capacity after HL stress compared with the wild type (Fig. 4A). Based on this observation, different photoprotective mechanisms, both nonenzymatic and enzymatic, that are normally activated during HL stress were investigated in more detail. Already under control conditions, total carotenoid content was 10% to 15% lower in cytokinin-deficient plants in comparison with wild-type plants. This difference was even more pronounced after HL treatment. The increase of the total carotenoid content shown in the wild type was almost completely absent in cytokinin-deficient plants (Fig. 4B). Among the carotenoids, the xanthophylls are of particular interest due to their involvement in energy dissipation during light stress. The amount of neoxanthin and zeaxanthin was already significantly decreased under control conditions in both ahk2 ahk3 and 35S:CKX4 plants. After HL treatment, the concentrations of all xanthophylls were more strongly increased in the wild type compared with plants with a reduced cytokinin status, pointing to a compromised energy dissipation through the xanthophyll cycle (Fig. 4C).
Figure 4.
Antioxidant capacity in plants with a reduced cytokinin status after HL treatment. Detached leaves (pooled leaves 5, 6, and 7) of 4-week-old plants were exposed to HL for 24 h. A, Total antioxidant capacity of detached leaves expressed as ORAC units (µmol Trolox equivalents g−1 fresh weight). B to D, Lipophilic antioxidant contents. Total carotenoid content (B), content of xanthophyll pigments (C; NEO, neoxanthin; VIO, violaxanthin; ANTH, antheraxanthin; ZEA, zeaxanthin), and tocopherol content (D) are shown. E and F, Hydrophilic antioxidant contents. Ascorbic acid (E; ASC, ascorbate; DHA, dehydroascorbate) and glutathione (F; GSH, reduced glutathione; GSSG, oxidized glutathione) contents are shown. Asterisks indicate significant differences from the respective wild-type (WT) value (* 0.05 > P > 0.01; ** 0.01 > P > 0.001; *** P < 0.001). Error bars represent se (n = 4 in A–C; n = 12 in D–F). FW, Fresh weight.
Furthermore, we also evaluated the tocopherol (Fig. 4D), ascorbate (Fig. 4E), and glutathione (Fig. 4F) contents. The total tocopherol content was increased in ahk2 ahk3 and 35S:CKX4 plants after HL treatment above the wild-type level, which mainly resulted from the elevation in α-tocopherol. Ascorbate levels increased as well after HL treatment, but the increase was similar in both cytokinin-deficient and wild-type plants. Total glutathione levels increased only slightly after HL exposure, while especially the oxidized form was highest in 35S:CKX4 plants (Fig. 4F).
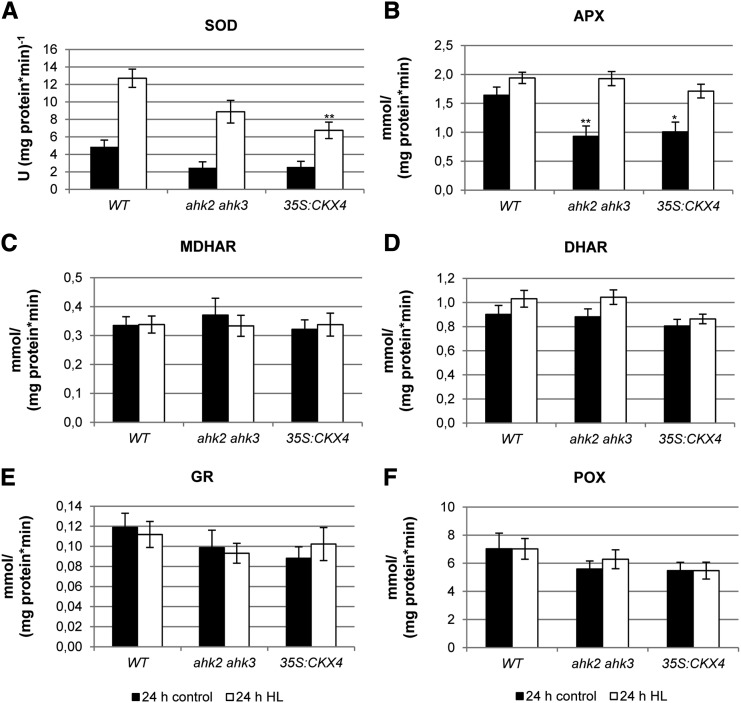
The enzyme activities of SOD and of the Halliwell-Asada pathway, including APX, monodehydroascorbic acid reductase (MDHAR), dehydroascorbic acid reductase (DHAR), and glutathione reductase (GR), as well as the activity of peroxidase (POX; Fig. 5) were analyzed in more detail. Only SOD and APX activities were altered upon HL exposure and showed significant differences between the genotypes (Fig. 5, A and B). Interestingly, ahk2 ahk3 and 35S:CKX4 plants showed a strong reduction of APX and SOD activities (by about 2-fold) under control conditions. After HL treatment, the APX activity increased only slightly in wild-type plants, while in both ahk2 ahk3 and 35S:CKX4 plants, there was a strong increase (2- and 1.7-fold, respectively). The SOD activities in HL-treated wild-type and 35S:CKX4 plants increased by 2.5-fold, and ahk2 ahk3 mutants had even a 3.5-fold increase in activity. Yet, the SOD activity in plants with a reduced cytokinin status remained significantly lower compared with wild-type plants (Fig. 5A).
Figure 5.
Scavenging enzyme activities in plants with a reduced cytokinin status after HL treatment. Detached leaves (pooled leaves 5, 6, and 7) of 4-week-old Arabidopsis plants were exposed for 24 h to HL. A, SOD. B, APX. C, MDHAR. D, DHAR. E, GR. F, POX. Asterisks indicate significant differences from the respective wild-type (WT) value (* 0.05 > P > 0.01; ** 0.01 > P > 0.001). Error bars represent se (n = 12). U, Units.
Alterations in Chloroplast Ultrastructure in Cytokinin-Deficient Plants
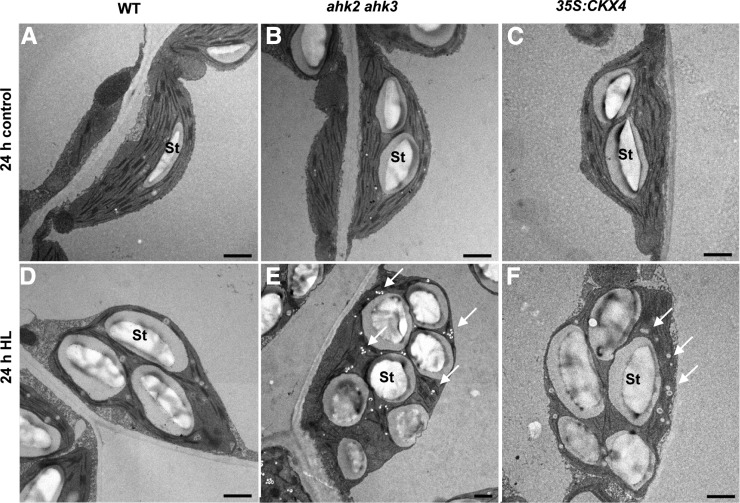
In order to explore whether the higher sensitivity to light stress of plants with a reduced cytokinin status would result in changes in chloroplast ultrastructure, we analyzed chloroplasts in control and HL-treated leaves by transmission electron microscopy. Already under control conditions, chloroplasts of ahk2 ahk3 and 35S:CKX4 plants showed ultrastructural differences compared with the wild type. Chloroplasts were not lens-shaped, as in wild-type plants, but slightly swollen, probably due to the presence of large starch grains. No alterations were observed in thylakoids or in grana stacking (Fig. 6, A–C). Following HL treatment, chloroplasts of all genotypes contained reduced size of grana stacks, large starch grains, and many and large plastoglobuli. However, chloroplasts of plants with a reduced cytokinin status showed a stronger increase in the number of plastoglobuli and starch grains (Fig. 6, E and F, arrows).
Figure 6.
Influence of HL on the chloroplast ultrastructure in the wild type (WT) and plants with a reduced cytokinin status. Images show chloroplasts in detached leaves (leaf 6) of 4-week-old Arabidopsis plants following exposure for 24 h to HL. Note the increased starch (St) content and the number of plastoglobuli (indicated by arrows) in plants with a reduced cytokinin status. Bars = 1,200 nm.
Cytokinin Signaling Genes Are Required for the Light Stress Response
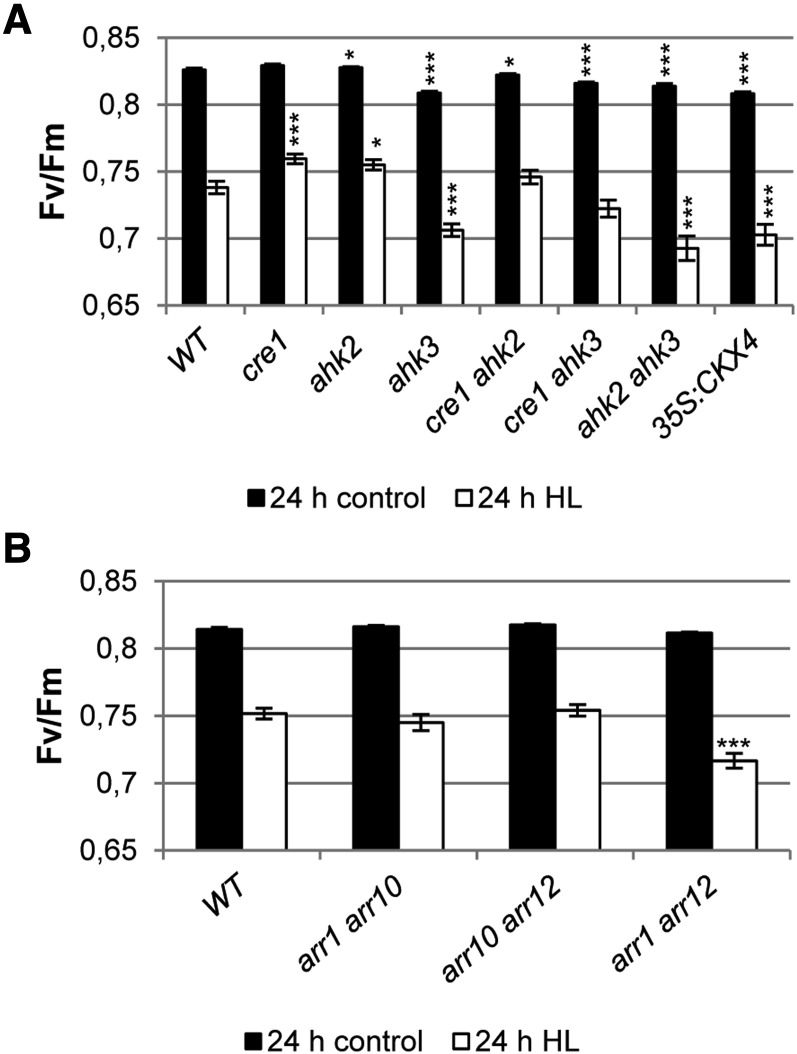
To study the contribution of the different cytokinin receptors in mediating the light stress response, we analyzed all single (cre1, ahk2, and ahk3) and double (cre1 ahk2, cre1 ahk3, and ahk2 ahk3) cytokinin receptor mutants after 24 h of HL (Fig. 7A). Mutations of AHK2 and/or CRE1/AHK4 alone did not cause a decreased activity of PSII (Fv/Fm) compared with the wild type, while a significant decrease in PSII activity was observed in ahk3 mutants. The decrease in Fv/Fm was even a bit stronger in the ahk2 ahk3 double receptor mutant, which could not be observed in the other double receptor mutants. This indicates that the AHK3 receptor is the key mediator in this light stress response, while AHK2 probably has an accessory function.
Figure 7.
The cytokinin receptors AHK2 and AHK3 and the type B response regulators ARR1 and ARR12 mediate the cytokinin-dependent light stress response. Soil-grown 4-week-old plants were continuously grown under SD conditions or exposed for 24 h to HL. A, Fv/Fm in detached leaf 6 of different cytokinin receptor single and double mutants after 24 h of HL (n = 15). B, Fv/Fm in detached leaf 6 of different B-type response regulator double mutants (n = 10). Asterisks indicate significant differences compared with the wild type (WT) (* 0.05 > P > 0.01; *** P < 0.001). Error bars represent se.
Subsequently, we investigated the possible involvement of type B ARRs. We chose to analyze loss-of-function mutants of the B-type ARR genes ARR1, ARR10, and ARR12, because mutant analysis had shown that the corresponding transcription factors mediate the majority of cytokinin action in different processes during vegetative development (Argyros et al., 2008; Ishida et al., 2008). Fv/Fm values were measured after HL treatment in several double mutants (arr1 arr10, arr1 arr12, and arr10 arr12; Fig. 7B). A significantly decreased PSII activity was observed in arr1 arr12 mutants, while no difference from the wild type was noted for the other two double mutants investigated. This demonstrates that ARR1 and ARR12 act redundantly downstream of AHK3 and AHK2 in mediating the cytokinin-regulated light stress response.
DISCUSSION
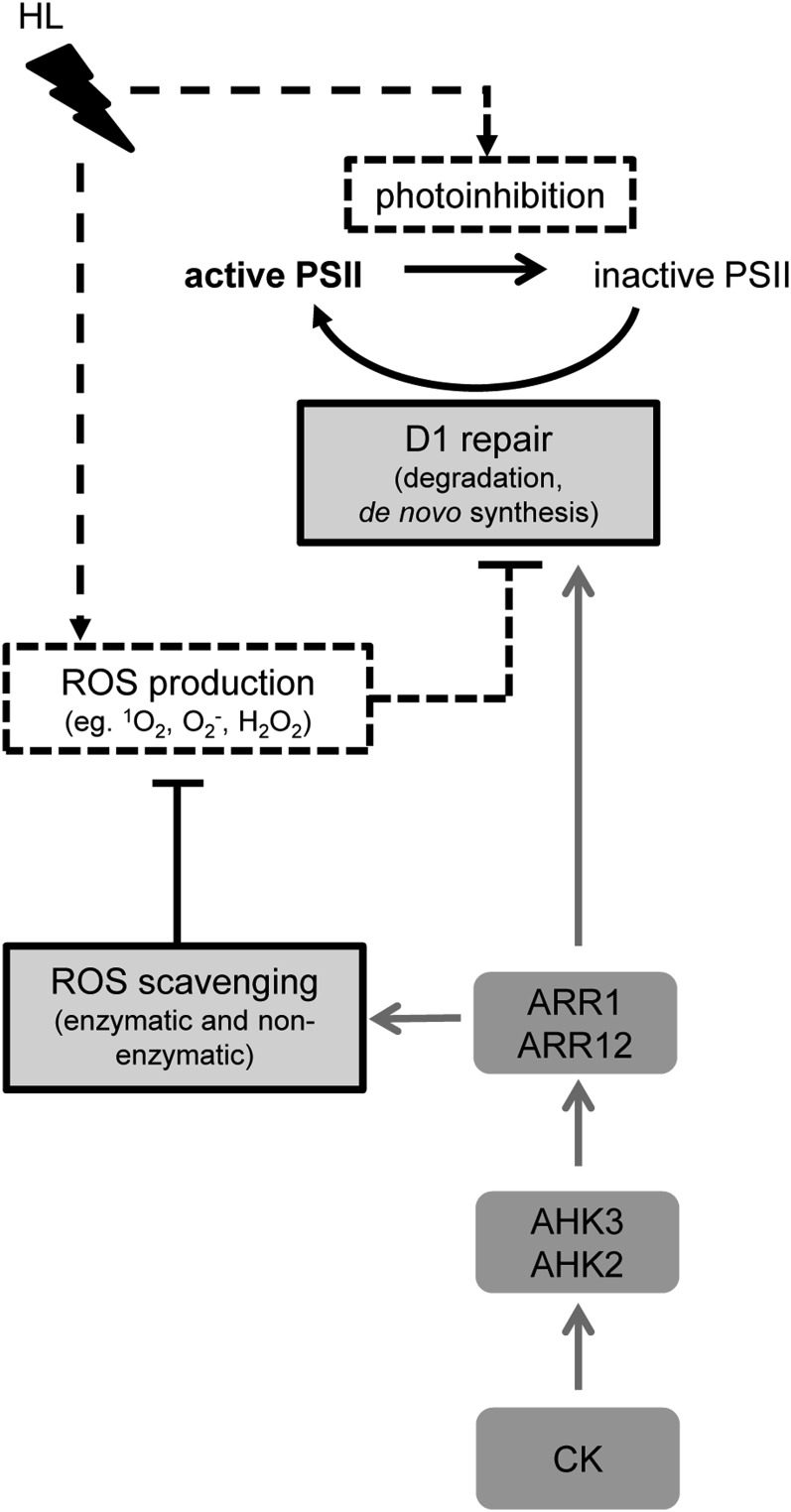
In this study, we have investigated the role of cytokinin during HL stress by evaluating the responses of Arabidopsis plants with a reduced cytokinin signaling or content. HL stress caused stronger damage to the photosynthetic apparatus in plants with a lower cytokinin status, which was apparent as a reduced Fv/Fm (Fig. 1) and a lower capability of these plants to recover from photoinhibition after HL stress (Fig. 2A). The D1 protein level was strongly reduced in cytokinin-deficient plants, reflecting severe photoinhibition of PSII (Fig. 2B). The slower and incomplete recovery from light stress in cytokinin-deficient plants indicates insufficient repair of PSII, which could be either due to a compromised D1 repair machinery and/or to a persistent or even irreversible photodamage. Further research into the mechanisms revealed that the ROS-scavenging capacity was significantly diminished in cytokinin-deficient plants (Figs. 4 and 5), which contributed to their higher susceptibility. The cytokinin-dependent light stress responses are mainly mediated by the cytokinin receptor AHK3 (and to a lesser extent by AHK2) and the B-type response regulators ARR1 and ARR12, as summarized in a model (Fig. 8).
Figure 8.
Model for the protective function of cytokinin in the light stress response. HL causes the inhibition of the active PSII and the production of ROS, which can also inhibit D1 repair (dotted black lines). Plants have evolved several mechanisms, such as ROS scavenging and the D1 repair cycle, to counteract the destructive effect of too much light (light gray boxes and solid black lines). Cytokinin (CK) has protective functions in the light stress response by promoting both ROS scavenging and D1 repair (gray lines), which is mediated by the AHK2 and AHK3 receptors and the type B response regulators ARR1 and ARR12 acting downstream of the receptors. The involvement of ARR1 and ARR12 has been concluded based on the results shown in Figure 7B.
Imbalance between Photodamage and Repair Causes the Cytokinin-Regulated Light Stress Response
The extent of photoinhibition is associated with a balance between the rate of photodamage and its repair (Takahashi and Badger, 2011). One indication for an impaired D1 repair cycle in cytokinin-deficient plants was their reduced capacity to recover from photoinhibition after relaxation (Fig. 2A). Moreover, the protein level of D1 was earlier and more strongly reduced following HL treatment in these plants in comparison with the wild type (Fig. 2B), suggesting an accelerated depletion of D1 (i.e. increased photodamage) and/or a lack of replenishment of the D1 protein pool by de novo synthesis. The lower transcript level of the D1 protein-encoding PSBA gene in cytokinin-deficient plants (Fig. 2C) hints at a possibly impaired D1 de novo synthesis. However, it should be noted that posttranscriptional mechanisms, including mRNA processing and cotranslational modifications, are the major steps in the regulatory network controlling the expression of the PSBA gene and the production of the D1 protein (Mulo et al., 2012). These mechanisms could also be affected in plants with a reduced cytokinin status.
There are several important steps in the degradation of damaged and the maturation of novel D1 protein. ATP-dependent FTSH metalloproteases and ATP-independent DEG endopeptidases play predominant roles in D1 degradation (Kato and Sakamoto, 2009). Arabidopsis mutants of the major isoforms of FTSH (FTSH2 and FTSH5) showed a high sensitivity to photoinhibition under HL and accumulated high levels of ROS (Sakamoto et al., 2004), and deg5 deg8 mutants also exhibited an HL-sensitive phenotype (Sun et al., 2007). These reports clearly demonstrate that a proper degradation of the damaged D1 is important for protection against photoinhibition. At the end of the D1 repair cycle, newly synthesized preD1 needs to be processed to mature D1 by CTP activity (Anbudurai et al., 1994; Roose and Pakrasi, 2004). Recently, mutant analysis has identified one of the three predicted Arabidopsis CTP homologs (At4g17740) to be required for an efficient repair of D1 under HL (Che et al., 2013). Our experiments showed that HL caused no major differences in the expression of genes encoding FTSH and DEG proteases between wild-type and cytokinin-deficient plants (Fig. 2, D and E). Also, the functionally important CTP gene homolog (At4g17740) was neither induced by HL nor differently regulated among the investigated genotypes. One other CTP homolog (At3g57680) exhibited a different expression pattern in cytokinin-deficient plants compared with the wild type (Fig. 2F). However, the corresponding loss-of-function mutant (Yin et al., 2008) showed a wild-type-like HL response under our conditions. This indicates that At3g57680 is not indispensable under HL stress, which is consistent with the report of Che et al. (2013). Together, the transcript data suggest that cytokinin does not act mainly through transcriptional regulation of the analyzed genes but may act on a different level to influence the efficiency of the D1 repair cycle.
An important result to understand the light stress phenotype of cytokinin-deficient plants is the stronger reduction of D1 protein as a consequence of increased photodamage and an impaired D1 repair cycle, which is a major cause for photoinhibition in these plants upon exposure to HL. The available data do not allow us to distinguish whether (1) limited ROS scavenging causing enhanced ROS production and thereby indirectly inhibiting the D1 repair cycle (Fig. 8), (2) reduced de novo production of the D1 protein, or (3) an impaired action of proteins required for a functional D1 repair cycle contribute most to the reduction and impaired recovery of the D1 protein.
Cytokinin Deficiency Causes a Reduced Antioxidant Capacity
An initial experiment showed that in plants with a reduced cytokinin status, the total antioxidant capacity is decreased in comparison with the wild type. Further investigation revealed that this reduction is mostly due to a lower level of antioxidative compounds, such as carotenoids and, more specifically, xanthophylls (Fig. 4). In contrast, no major differences between wild-type and cytokinin-deficient plants were found for ascorbate and glutathione, which also play an important role in redox homeostasis (for review, see Foyer and Noctor, 2011). Carotenoids of the xanthophyll cycle are of great importance for the dissipation of excess excitation energy as quenchers of 3chl* and 1O2 (Demmig-Adams and Adams, 1996). The lower increase of these pigments in plants with a reduced cytokinin status in response to HL could result in a compromised xanthophyll cycle. Especially neoxanthin levels, which are known to be specifically involved in the scavenging of superoxide (Dall’Osto et al., 2007), were decreased in cytokinin-deficient plants under both control and stress conditions. In contrast, the content of tocopherols, which are lipid-soluble antioxidants scavenging ROS and nitrogen species, protecting membrane lipids from autocatalytic oxidation and peroxidation (Schneider, 2005; Krieger-Liszkay and Trebst, 2006), was increased. This increase was mainly due to an increase in α-tocopherol, which plays an important role in protecting PSII from 1O2 (Trebst et al., 2002).
The amount of plastoglobuli, storage sites for lipoprotein particles containing, for example, plastoquinone, α-tocopherol and triacylglycerols (Lundquist et al., 2012), and the number of starch granules, were strongly increased in cytokinin-deficient plants. A higher starch content was also observed in the sink leaves of transgenic tobacco (Nicotiana tabacum) plants expressing 35S:CKX1 and 35S:CKX2 (Werner et al., 2008). Of particular interest in the context of the HL stress phenotype is the larger number of plastoglobuli. Numerous studies reported a strong increase in plastoglobuli size and amount under stress conditions, which has been connected to the antioxidative effect of tocopherol (Bréhélin et al., 2007). This might also be the case in cytokinin-deficient plants, where the higher increase in tocopherol levels could be part of a compensatory protective mechanism counteracting a higher degree of photodamage. Compensatory processes have been described in several studies analyzing antioxidant protection against photoinhibition. For example, the lutein-deficient2-1 (lut2-1) Arabidopsis mutant, which is unable to synthesize lutein, compensates by increased amounts of violaxanthin (Dall’Osto et al., 2006). The non-photochemical quenching1 (npq1) npq4 lut2 mutant compensates for the thermal dissipation energy deficit with increased α-tocopherol and ascorbate levels (Golan et al., 2006), and α-tocopherol-deficient Arabidopsis mutant (vitamin e-deficient1) augmented the synthesis of zeaxanthin (Havaux et al., 2005).
Recycling enzymes of the Halliwell-Asada pathway (MDHAR, DHAR, and GR) showed no altered activity in response to HL, while the enzymes directly involved in scavenging of superoxide (SOD) and H2O2 (APX) were strongly activated upon HL (Fig. 5). Cytokinin-deficient plants showed only about half of the SOD activity of wild-type plants under control conditions and after HL treatment (Fig. 5A). A similar reduction in activity was also noted for APX under control conditions, but this was compensated upon HL treatment. These results indicate, on the one hand, that the scavenging capacity of cytokinin-deficient plants is generally lower than in wild-type plants and, on the other hand, that upon HL they seem to encounter more oxidative stress and try to deal with it by increasing the activities of APX and SOD.
AHK2 and AHK3 Mediate the Cytokinin Function in the Light Stress Response Acting through ARR1 and ARR12
A strong HL effect was caused by the loss of AHK3 and was further enhanced by the additional loss of AHK2, which alone was ineffective. This indicates that AHK3 has a major role in the HL response while AHK2 has a cooperative function. Similar observations on cooperative or redundant functions of AHK2 and AHK3 have been made in other studies (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Interestingly, AHK2 and AHK3 are evolutionarily more closely related to each other than to CRE1/AHK4, and both receptors are predominantly expressed and active in shoot tissues (Ueguchi et al., 2001; Higuchi et al., 2004; Stolz et al., 2011). Several important functions during leaf development have been attributed to AHK2 and AHK3, including a role in leaf cell formation, chlorophyll metabolism, and leaf senescence (Kim et al., 2006; Riefler et al., 2006). However, a role in the light stress response has not yet been listed among their various activities (Müller, 2011; Heyl et al., 2012). An apparently higher relevance of AHK3 compared with AHK2 could be due to the different sensitivities of the receptors to various cytokinin metabolites (Spíchal et al., 2004; Romanov et al., 2006; Stolz et al., 2011) and/or to differences in coupling to downstream signaling elements, as indicated by distinct interacting proteins (Dortay et al., 2008).
The fact that the arr1 arr12 mutant also displayed a strong HL effect indicates that cytokinin action in the light stress response is mediated at least partly through transcriptional regulation. ARR1 and ARR12 belong to subfamily 1 of the type B response regulators (Heyl et al., 2008; Hill et al., 2013), and the corresponding genes are expressed in leaves (Mason et al., 2004). Mutation of both genes was required to obtain an effect indicating their redundant action, which is a common feature of type B response regulators (Müller, 2011). For ARR1 and ARR12, a combined action in regulating the expression of HIGH-AFFINITY K+ TRANSPORTER1 (HKT1;1), encoding a sodium transporter, and the accumulation of sodium in Arabidopsis shoots has been reported (Mason et al., 2010). Interestingly, ARR10, which is the closest relative of ARR12 and also has a function in leaf development (Argyros et al., 2008; Ishida et al., 2008), is not required for the light stress response, indicating that it has functionally diverged.
The role of ARR1 and ARR12 in light stress protection links this activity to a specific transcriptional response, consistent with the great importance of a fine-tuned gene regulation to realize the many different biological activities of cytokinin (for review, see Brenner et al., 2012). Cytokinin regulates numerous genes involved in light signaling and redox regulation (Rashotte et al., 2003; Brenner et al., 2005; Taniguchi et al., 2007; Brenner and Schmülling 2012; Bhargava et al., 2013). For example, microarray meta-analyses placed genes encoding glutaredoxins, which have a role in protecting plants against photooxidative stress (Laporte et al., 2012), among the top 20 cytokinin-regulated genes (Brenner et al., 2012; Bhargava et al., 2013). The emerging network of transcriptional regulation connecting the actions of light, ROS, and cytokinin (Vandenbussche et al., 2007; Chen et al., 2013) provides a starting point to unravel the molecular mechanisms linking cytokinin with its function in light stress protection.
MATERIALS AND METHODS
Plant Material, Growth, and Treatment Conditions
Arabidopsis (Arabidopsis thaliana Columbia-0) wild-type plants, cytokinin-deficient 35S:CKX4 (Werner et al., 2003), cytokinin receptor mutant plants (ahk2-5 ahk3-7, cre1-2 ahk2-5, and cre1-2 ahk3-7; Riefler et al., 2006), B-type arr mutants (arr1-3 arr10-5, arr1-3 arr12-1, and arr10-5 arr12-1; Mason et al., 2004; Ishida et al., 2008), and the ctpa1 mutant (Yin et al., 2008) were grown in a growth chamber under controlled conditions: 150 to 200 µmol m−2 s−1, 8/16-h photoperiod (SD), 22°C, and 60% relative humidity. HL experiments (approximately 1,000 µmol m−2 s−1) were performed with detached leaves of 4-week-old plants floating on water or on whole plants in a growth cabinet (Percival AR66L; Percival Scientific) at 26°C and 60% relative humidity.
INCYDE Treatment
Five days before HL treatment, 4-week-old plants were sprayed once per day with a solution containing 10 µm INCYDE (Zatloukal et al., 2008) and 0.01% (v/v) Silwet L-77. Mock-treated samples were sprayed with a solution containing only Silwet L-77. Whole plants were exposed to the HL standard treatment described above.
Lincomycin Treatment
Detached leaves were incubated with their petioles in 1 mm lincomycin or water (control) for 3 h at 20 µmol m−2 s−1 (22°C) to block protein synthesis prior to photoinhibitory treatment. Incubated leaves were then illuminated at a photon flux density of 1,000 µmol m−2 s−1 (22°C) for 3 h before measurement of photochemical efficiency.
Fluorometry
Chlorophyll fluorescence emission was measured on detached leaves and defined leaves of whole plants with a modulated chlorophyll fluorometer (Photosystem Instruments). The maximal photochemical efficiency of PSII was determined after dark adaptation for 20 min from the ratio of variable (Fv) to maximum (Fm) fluorescence [Fv/Fm = (Fm − F0)/Fm]. An actinic light pulse (0.2 µmol m−2 s−1) was used to determine the initial (minimum) PSII fluorescence in the dark-adapted state (F0), and Fm was determined by a saturating light pulse (1,500 µmol m−2 s−1).
Transmission Electron Microscopy
Parts of the sixth leaf were fixed for 3 d at 4°C using vacuum infiltration in 2% (v/v) paraformaldehyde and 2% (v/v) glutaraldehyde buffered in 50 mm cacodylate buffer with 50 mm NaCl. Samples were washed with 50 mm cacodylate buffer containing 50 mm NaCl and with 50 mm glycylglycin buffer containing 100 mm NaCl. Postfixation was performed in 1% (w/v) osmium tetroxide buffered in 50 mm cacodylate buffer containing 50 mm NaCl for 3 h. After washing with distilled water, leaf tissues were incubated for 1 h in 0.1% (w/v) tannic acid in 100 mm HEPES buffer, rinsed with water, and incubated overnight at 4°C in water. After staining in 2% (w/v) uranyl acetate for 1.5 h, fixed tissues were dehydrated and embedded in Spurr’s epoxy resin. Ultrathin sections (65 nm), obtained using a Leica Ultracut UCT ultramicrotome, were mounted on 0.7% (w/v) formvar-coated copper grids (200 mesh). The sections were contrasted with uranyl acetate (2% [w/v] in 50% ethanol) followed by lead citrate (4% [w/v] solution) and examined with a FEI Tecnai Spirit transmission electron microscope operated at 120 kV.
Protein Isolation and Protein Gel-Blot Analysis
Frozen leaf material (pooled leaves 5, 6, and 7 from four different plants) was homogenized with the Retsch Mixer Mill MM2000 with two stainless-steel beads (2 mm diameter) in each sample. Total proteins were extracted from the homogenized leaf material in a buffer containing 150 mm NaCl, 100 mm Tris, and 1% Triton X-100 (pH 7). Protein concentrations were determined according to Bradford (1976). Proteins (3.5 µg) were separated by SDS-PAGE (14%) and blotted to a polyvinylidene difluoride membrane overnight at 4°C. Prior to blocking, blots were stained with Ponceau S (Sigma-Aldrich) to check correct loading, followed by washing three times for 5 min each in Tris-buffered saline plus 0.05% Tween 20 at room temperature. The blots were blocked for 1.5 to 2 h in 7% skim milk (in Tris-buffered saline plus 0.05% Tween 20; Sigma-Aldrich) at room temperature. Afterward, the blots were incubated for 1 h at room temperature with the primary antibody (polyclonal D1 specific; Agrisera) followed by the secondary antibody (goat anti-rabbit IgG peroxidase conjugate; Calbiochem). Both antibodies were diluted 1:30,000 in the blocking solution. After each antibody treatment, blots were washed as described above. Afterward, the D1 protein was immunodetected using the SuperSignal West Pico Chemiluminescent Substrate Kit (Fisher Scientific) according to the manufacturer’s instructions. Quantification of independent experiments (n = 3 [Fig. 3B] and n = 4 [Fig. 4C]) was performed with ImageJ 1.46r (National Institutes of Health).
Analysis of Transcript Levels by Quantitative Reverse Transcription-PCR
Total RNA was extracted from leaves (pooled leaves 5, 6, and 7 from four different plants) with the TRIzol method. TRIzol reagent (38% phenol, 0.8 m guanidinium thiocyanate, 0.4 m ammonium thiocyanate, 0.1 m sodium acetate, pH 5, and 5% glycerol) was made as described in the GIBCO TRIzol manual of Invitrogen. RNA was precipitated by the modified salt precipitation method for proteoglycan and polysaccharide contamination as described in the GIBCO troubleshooting guide. Total RNA was further purified using RNeasy mini-columns including the on-column DNase digestion as described in appendix D of the Qiagen RNeasy Mini Handbook. The purified RNA concentration was determined spectrophotometrically at 260 nm using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies). The RNA purity was evaluated by means of the 260:280 ratio. Equal amounts of starting material (1 µg of RNA) were used in a 10-µL SuperScript III Reverse Transcriptase reaction. First-strand complementary DNA synthesis was primed with a combination of oligo(dT) primers and random hexamers. Primer pairs were designed, using Primer 3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi), under the following conditions: optimum melting temperature at 60°C, GC content between 20% and 80%, 150-bp maximum length. Primers used for reference genes and genes of interest are listed in Supplemental Table S1. Real-time PCR using FAST SYBR Green I technology was performed on an ABI PRISM 7500 sequence detection system (Applied Biosystems) and universal “FAST” cycling conditions (10 min at 95°C, 40 cycles of 15 s at 95°C and 60 s at 60°C) followed by the generation of a dissociation curve to check for specificity of the amplification. Reactions contained SYBR Green Master Mix (Applied Biosystems), 300 nm of gene-specific forward and reverse primers, and 2 µL of the diluted complementary DNA in a 20-µL reaction. Gene expression data were normalized against three different nucleus-encoded reference genes (ACTIN2, KORRIGAN1, and TBP-ASSOCIATED FACTOR II15) or plastid-encoded reference genes (RESISTANCE TO PSEUDOMONAS SYRINGAE3 and NAD(P)H DEHYDROGENASE COMPLEX I) according to Vandesompele et al. (2002) and are presented relative to the control treatment.
ORAC Assay
The ORAC assay was carried out as described by Gillespie et al. (2007). Briefly, fluorescein (60 nm), antioxidant (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid [12.5 mm]; Trolox), and 2,2′-azobis-2-methyl-propanimidamide-dihydrochloride (AAHP [18.75 mm]) were incubated in 200 µL of KH2PO4-K2HPO4 buffer (75 mm; pH 7.0) at 37°C on a 96-well plate (Costar; Sigma-Aldrich). The fluorescence (excitation wavelength, 485 nm; emission wavelength, 520 nm) was monitored every 1 min over a period of 60 min by Synergy HT (BioTek).
Pigment Analysis
Chlorophyll and carotenoids were extracted and analyzed by HPLC applying the method of Thayer and Björkman (1990).
Glutathione and Ascorbic Acid Determination
Plant material (approximately 200 mg of pooled leaves 5, 6, and 7 from four different plants) was quickly ground in a MagNa Lyzer Instrument (Roche). Samples were kept frozen during grinding to prevent oxidation. Ice-cold 6% (w/v) meta-phosphoric acid was added, and samples were thawed on ice and centrifuged at 12,000g at 4°C for 15 min. The resulting supernatant was kept on ice until HPLC analysis. Antioxidants were separated on a 100-mm × 4.6-mm Polaris C18-A reverse-phase HPLC column (3-μm particle size, 40°C; Varian) with an isocratic flow of 1 mL min−1 of the elution medium (2 mm KCl, adjusted to pH 2.5 by o-phosphoric acid). The components were quantified using a homemade electrochemical detector with glassy carbon electrode and a Schott pt 62 reference electrode. The purity and identity of the peaks were confirmed using a diode-array detector (SPD-M10AVP; Shimadzu), which was placed in line with the electrochemical detector. The amount of oxidized dehydroascorbate or oxidized glutathione was measured indirectly as the difference between the total concentrations of antioxidants in a dithiothreitol-reduced fraction and the concentrations in the sample prior to reduction. Reduction of the sample was obtained by incubation of an aliquot of the extract in 400 mm Tris and 200 mm dithiothreitol for 10 min in the dark. The pH of this mixture was checked to be between 6 and 7. After 15 min, the pH was lowered again by 4-fold dilution in elution buffer prior to HPLC analysis.
Tocopherol Determination
Plant material (approximately 200 mg of pooled leaves 5, 6, and 7 from four different plants) was quickly ground in a MagNa Lyzer Instrument. Dimethyltocol (5 μg mL−1) was added to each sample as an internal standard. Hexane was used to extract tocopherols, and after centrifugation and filtration, tocopherols were separated on a Partisil Pac 250-mm × 4.6-mm column (5-μm particle size, length of 250 mm, i.d. of 4.6 mm; Shimadzu) with an isocratic flow of 1.5 mL min−1 of the elution buffer (hexane containing 8% tetrahydrofuran). Fluorescence was detected with a Shimadzu spectrofluorometric detector RF-10A (excitation wavelength, 290 nm; emission wavelength, 330 nm). Data were analyzed with Shimadzu Class VP 6.14 software.
Analysis of Enzyme Activities
Activities of APX (EC 1.11.1.11), DHAR (EC 1.8.5.1), MDHAR (EC1.6.5.4), GR (EC 1.8.1.7), and POX (EC 1.11.1) were measured as described by Murshed et al. (2008) with slight modifications. Briefly, frozen leaf tissue (approximately 150 mg of pooled leaves 5, 6, and 7 from four different plants) was homogenized with the Retsch Mixer Mill MM2000 with two stainless-steel beads (2 mm diameter) in each sample. Extraction of the enzymes was performed in 1 mL of ice-cold 50 mm MES-KOH buffer (pH 6.0) containing 40 mm KCl, 2 mm CaCl2, and 1 mm l-ascorbic acid followed by vortexing and centrifugation at 16,000g for 20 min at 4°C. All enzyme assays were performed in a final volume of 0.2 mL on 96-well microtiter plates at 25°C (PowerWave HT microplate spectrophotometer; BioTek). Samples and blanks were analyzed in triplicate. The activity of SOD (EC 1.15.1.1) was determined by measuring its ability to inhibit the photochemical reduction of nitroblue tetrazolium using the method of Beauchamp and Fridovich (1973). The 1-mL reaction mixture contained a 50 mm potassium phosphate buffer (pH 7.8) with 0.933 m Met, 75 µm nitroblue tetrazolium, 0.1 mm EDTA, 10 µm riboflavin, and 30 µL of leaf extract. The mixture was vortexed and divided into wells of a 96-well plate (200 µL per well). The reaction was started by putting the plate into a light chamber (Percival AR66; illumination at 1,000 µmol photons m−2 s−1) for 2 min. Absorbance of the reaction mixture at 550 nm was read. A nonirradiated reaction mixture served as a control. SOD activity was calculated based on a SOD standard curve.
Statistical Analysis
Statistical analyses were performed using SAS version 9.1.3 (ANOVA, Tukey’s post hoc test). Normality and homogeneity of variance were tested using the Shapiro-Wilk and Levene tests (Neter et al., 1996). In order to meet the assumptions, data sets were transformed using log or square-root transformation. All data sets, except fluorometry data, fulfilled the assumptions for ANOVA. If assumptions were not met, a nonparametric Kruskal-Wallis test was performed followed by a Mann-Whitney test to perform a pairwise comparison.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primer sequences used for quantitative real-time PCR.
Acknowledgments
We thank Rudi Lurz and Beatrix Fauler (Max Planck Institute for Molecular Genetics) for technical support with electron microscopy.
Glossary
- HL
high light
- ROS
reactive oxygen species
- 1O2
singlet oxygen
- H2O2
hydrogen peroxide
- 3chl*
triplet chlorophyll
- SOD
superoxide dismutase
- APX
ascorbate peroxidase
- SD
short-day
- Fv/Fm
ratio of variable fluorescence to maximum fluorescence
- ORAC
oxygen radical antioxidant capacity
- MDHAR
monodehydroascorbic acid reductase
- DHAR
dehydroascorbic acid reductase
- GR
glutathione reductase
- POX
peroxidase
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. Schm 814/27–1 and CRC 973).
These authors contributed equally to the article.
The online version of this article contains Web-only data.
References
- Adir N, Zer H, Shochat S, Ohad I. (2003) Photoinhibition: a historical perspective. Photosynth Res 76: 343–370 [DOI] [PubMed] [Google Scholar]
- Anbudurai PR, Mor TS, Ohad I, Shestakov SV, Pakrasi HB. (1994) The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc Natl Acad Sci USA 91: 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Argueso CT, Ferreira FJ, Kieber JJ. (2009) Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ 32: 1147–1160 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro EM, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamäki E. (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56: 347–356 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Asada K. (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Aro EM. (2002) Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond B Biol Sci 357: 1451–1459, discussion 1459–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber J, Andersson B. (1992) Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci 17: 61–66 [DOI] [PubMed] [Google Scholar]
- Beauchamp CO, Fridovich I. (1973) Isozymes of superoxide dismutase from wheat germ. Biochim Biophys Acta 317: 50–64 [DOI] [PubMed] [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang YH, Schaller GE, Loraine A, Kieber JJ. (2013) Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol 162: 272–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonman A, Prinsen E, Gilmer F, Schurr U, Peeters AJ, Voesenek LA, Pons TL. (2007) Cytokinin import rate as a signal for photosynthetic acclimation to canopy light gradients. Plant Physiol 143: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Analytical Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bréhélin C, Kessler K, van Wijk KJ. (2007) Plastoglobules: versatile lipoprotein particles in plastids. Trends Plant Sci 12: 260–266 [DOI] [PubMed] [Google Scholar]
- Brenner W, Schmülling T. (2012) Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific cytokinin responses. BMC Plant Biol 12: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T. (2012) Gene regulation by cytokinin in Arabidopsis. Front Plant Sci 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, Morelli G, Ruberti I. (2007) Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev 21: 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catský J, Pospíšilová J, Macháčková I, Wilhelmová N, Šesták Z. (1993) Photosynthesis and water relations in transgenic tobacco plants with T-DNA carrying gene 4 for cytokinin synthesis. Biol Plant 104: 339–347 [Google Scholar]
- Che Y, Fu A, Hou X, McDonald K, Buchanan BB, Huang W, Luan S. (2013) C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc Natl Acad Sci USA 110: 16247–16252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R. (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherniad’ev II. (2000) [Ontogenetic changes in the photosynthetic apparatus and effect of cytokinins]. Prikl Biokhim Mikrobiol 36: 611–625 [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M. (1994) A role for cytokinins in de-etiolation in Arabidopsis: det mutants have an altered response to cytokinins. Plant Physiol 104: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A, Valcke R. (2012) Evaluation of the photosynthetic activity in transgenic tobacco plants with altered endogenous cytokinin content: lessons from cytokinin. Physiol Plant 144: 394–408 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L, Cazzaniga S, North H, Marion-Poll A, Bassi R. (2007) The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. Plant Cell 19: 1048–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L, Lico C, Alric J, Giuliano G, Havaux M, Bassi R. (2006) Lutein is needed for efficient chlorophyll triplet quenching in the major LHCII antenna complex of higher plants and effective photoprotection in vivo under strong light. BMC Plant Biol 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW. (1996) The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci 1: 21–26 [Google Scholar]
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmülling T, Heyl A. (2008) Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J Proteome Res 7: 3649–3660 [DOI] [PubMed] [Google Scholar]
- Edelman M, Mattoo AK. (2008) D1-protein dynamics in photosystem II: the lingering enigma. Photosynth Res 98: 609–620 [DOI] [PubMed] [Google Scholar]
- Fischer BB, Hideg E, Krieger-Liszkay A. (2013) Production, detection, and signaling of singlet oxygen in photosynthetic organisms. Antioxid Redox Signal 18: 2145–2162 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie KM, Chae JM, Ainsworth EA. (2007) Rapid measurement of total antioxidant capacity in plants. Nat Protoc 2: 867–870 [DOI] [PubMed] [Google Scholar]
- Golan T, Müller-Moulé P, Niyogi KK. (2006) Photoprotection mutants of Arabidopsis thaliana acclimate to high light by increasing photosynthesis and specific antioxidants. Plant Cell Environ 29: 879–887 [DOI] [PubMed] [Google Scholar]
- Grennan AK, Ort DR. (2007) Cool temperatures interfere with D1 synthesis in tomato by causing ribosomal pausing. Photosynth Res 94: 375–385 [DOI] [PubMed] [Google Scholar]
- Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. (2012) Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci 17: 172–179 [DOI] [PubMed] [Google Scholar]
- Havaux M, Eymery F, Porfirova S, Rey P, Dörmann P. (2005) Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17: 3451–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Ramireddy E, Brenner WG, Riefler M, Allemeersch J, Schmülling T. (2008) The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol 147: 1380–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Riefler M, Romanov GA, Schmülling T. (2012) Properties, functions and evolution of cytokinin receptors. Eur J Cell Biol 91: 246–256 [DOI] [PubMed] [Google Scholar]
- Heyl A, Schmülling T. (2003) Cytokinin signal perception and transduction. Curr Opin Plant Biol 6: 480–488 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Mathews DE, Kim HJ, Street IH, Wildes SL, Chiang YH, Mason MG, Alonso JM, Ecker JR, Kieber JJ, et al. (2013) Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiol 162: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T. (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, Cho C, Lee DJ, Lee EJ, Strnad M, et al. (2010) A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem 285: 23371–23386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W. (2009) Protein quality control in chloroplasts: a current model of D1 protein degradation in the photosystem II repair cycle. J Biochem 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Trebst A. (2006) Tocopherol is the scavenger of singlet oxygen produced by the triplet states of chlorophyll in the PSII reaction centre. J Exp Bot 57: 1677–1684 [DOI] [PubMed] [Google Scholar]
- Kusnetsov VV, Herrmann RG, Kulaeva ON, Oelmüller R. (1998) Cytokinin stimulates and abscisic acid inhibits greening of etiolated Lupinus luteus cotyledons by affecting the expression of the light-sensitive protochlorophyllide oxidoreductase. Mol Gen Genet 259: 21–28 [DOI] [PubMed] [Google Scholar]
- Laporte D, Olate E, Salinas P, Salazar M, Jordana X, Holuigue L. (2012) Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J Exp Bot 63: 503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochmanová G, Zdráhal Z, Konecná H, Koukalová S, Malbeck J, Soucek P, Válková M, Kiran NS, Brzobohaty B. (2008) Cytokinin-induced photomorphogenesis in dark-grown Arabidopsis: a proteomic analysis. J Exp Bot 59: 3705–3719 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ. (2012) The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol 158: 1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macková H, Hronková M, Dobrá J, Turečková V, Novák O, Lubovská Z, Motyka V, Haisel D, Hájek T, Prášil IT, et al. (2013) Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J Exp Bot 64: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Schaller GE. (2010) Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J 64: 753–763 [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE. (2004) Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 135: 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty P, Allakhverdiev SI, Murata N. (2007) Application of low temperatures during photoinhibition allows characterization of individual steps in photodamage and the repair of photosystem II. Photosynth Res 94: 217–224 [DOI] [PubMed] [Google Scholar]
- Mok MC (1994) Cytokinins and plant development. In DWS Mok, Mok MC, eds, Cytokinins: Chemistry, Activity and Function. CRC Press, Boca Raton, FL, pp 57–79 [Google Scholar]
- Müller B. (2011) Generic signal-specific responses: cytokinin and context-dependent cellular responses. J Exp Bot 62: 3273–3288 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2007) Arabidopsis cytokinin signaling pathway. Sci STKE 2007: cm5. [DOI] [PubMed] [Google Scholar]
- Mulo P, Sakurai I, Aro EM. (2012) Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: From transcription to PSII repair. BBA - Bioenergetics 1817: 247–257 [DOI] [PubMed] [Google Scholar]
- Murshed R, Lopez-Lauri F, Sallanon H. (2008) Microplate quantification of enzymes of the plant ascorbate-glutathione cycle. Anal Biochem 383: 320–322 [DOI] [PubMed] [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W (1996) Applied Linear Statistic Models. McGraw-Hill, New York [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Kojima M, Werner T, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, Sakakibara H, Schmülling T, et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N. (2001) Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J 20: 5587–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthier B. (1979) The role of phytohormones (cytokinin) in chloroplasts development. Biochem Physiol Pflanz 174: 173–214 [Google Scholar]
- Procházková D, Haisel D, Wilhelmová N. (2008) Antioxidant protection during ageing and senescence in chloroplasts of tobacco with modulated life span. Cell Biochem Funct 26: 582–590 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Carson SDB, To JPC, Kieber JJ. (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132: 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104: 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN, Schmülling T. (2006) Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J Exp Bot 57: 4051–4058 [DOI] [PubMed] [Google Scholar]
- Roose JL, Pakrasi HB. (2004) Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J Biol Chem 279: 45417–45422 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Miura E, Kaji Y, Okuno T, Nishizono M, Ogura T. (2004) Allelic characterization of the leaf-variegated mutation var2 identifies the conserved amino acid residues of FtsH that are important for ATP hydrolysis and proteolysis. Plant Mol Biol 56: 705–716 [DOI] [PubMed] [Google Scholar]
- Satoh K, Yamamoto Y. (2007) The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynth Res 94: 203–215 [DOI] [PubMed] [Google Scholar]
- Schneider C. (2005) Chemistry and biology of vitamin E. Mol Nutr Food Res 49: 7–30 [DOI] [PubMed] [Google Scholar]
- Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov GA, Strnad M, Schmülling T. (2004) Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol 45: 1299–1305 [DOI] [PubMed] [Google Scholar]
- Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T. (2011) The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J 67: 157–168 [DOI] [PubMed] [Google Scholar]
- Sun X, Wang L, Zhang L. (2007) Involvement of DEG5 and DEG8 proteases in the turnover of the photosystem II reaction center D1 protein under heat stress in Arabidopsis thaliana. Chin Sci Bull 52: 1742–1745 [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. (2001) The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Synková H, Semorádová Š, Schnablová R, Witters E, Hŭsák M, Valcke R. (2006) Cytokinin-induced activity of antioxidant enzymes in transgenic Pssu-ipt tobacco during plant ontogeny. Biol Plant 50: 31–41 [Google Scholar]
- Synková H, Van Loven K, Pospíšilová J, Valcke R. (1999) Photosynthesis of transgenic Pssu-ipt tobacco. J Plant Physiol 155: 173–182 [Google Scholar]
- Takahashi S, Badger MR. (2011) Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci 16: 53–60 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Bauwe H, Badger M. (2007) Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol 144: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Murata N. (2008) How do environmental stresses accelerate photoinhibition? Trends Plant Sci 13: 178–182 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277 [DOI] [PubMed] [Google Scholar]
- Thayer SS, Björkman O. (1990) Leaf xanthophyll content and composition in sun and shade determined by HPLC. Photosynth Res 23: 331–343 [DOI] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebst A, Depka B, Holländer-Czytko H. (2002) A specific role for tocopherol and of chemical singlet oxygen quenchers in the maintenance of photosystem II structure and function in Chlamydomonas reinhardtii. FEBS Lett 516: 156–160 [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S. (2001) The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol 42: 751–755 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, Ahmad M. (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J 49: 428–441 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: H0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Holst K, Pörs Y, Guivarc’h A, Mustroph A, Chriqui D, Grimm B, Schmülling T. (2008) Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J Exp Bot 59: 2659–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T. (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Schmülling T. (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Aminaka R, Yoshioka M, Khatoon M, Komayama K, Takenaka D, Yamashita A, Nijo N, Inagawa K, Morita N, et al. (2008) Quality control of photosystem II: impact of light and heat stresses. Photosynth Res 98: 589–608 [DOI] [PubMed] [Google Scholar]
- Yaronskaya E, Vershilovskaya I, Poers Y, Alawady AE, Averina N, Grimm B. (2006) Cytokinin effects on tetrapyrrole biosynthesis and photosynthetic activity in barley seedlings. Planta 224: 700–709 [DOI] [PubMed] [Google Scholar]
- Yin Y, Sun X, Zhang L. (2008) An Arabidopsis ctpa homologue is involved in the repair of photosystem II under high light. Chin Sci Bull 53: 1021–1026 [Google Scholar]
- Zatloukal M, Gemrotová M, Dolezal K, Havlícek L, Spíchal L, Strnad M. (2008) Novel potent inhibitors of A. thaliana cytokinin oxidase/dehydrogenase. Bioorg Med Chem 16: 9268–9275 [DOI] [PubMed] [Google Scholar]
- Zubo YO, Selivankina SY, Yamburenko MV, Zubkova NK, Kulaeva ON, Kusnetsov VV. (2005) Cytokinins activate transcription of chloroplast genes. Dokl Biochem Biophys 400: 48–51 [DOI] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, Shakirova FM, Avalbaev AM, Kudryakova NV, Zubkova NK, Liere K, Kulaeva ON, Kusnetsov VV, et al. (2008) Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]