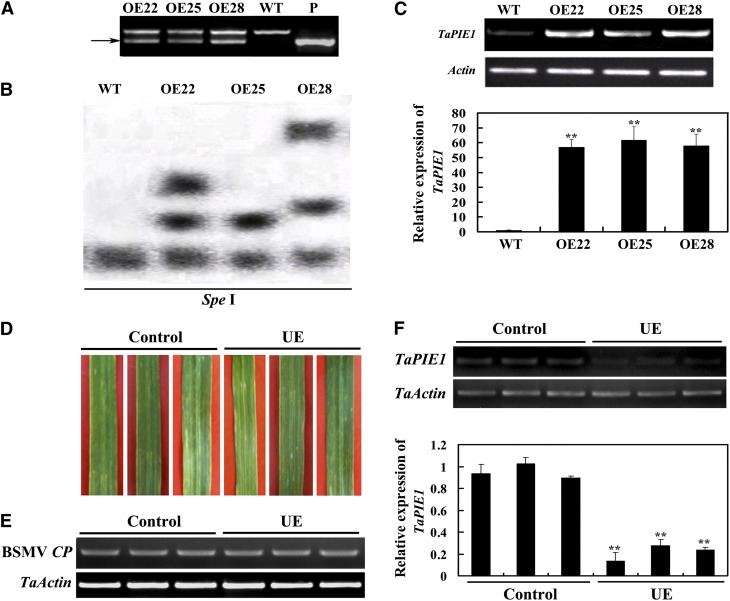
Figure 1.
Molecular characterization of TaPIE1-overexpressing transgenic wheat and TaPIE1-underexpressing wheat plants. A, PCR patterns of TaPIE1-overexpressing lines (OE; OE22, OE25, and OE28) and wild-type (WT) cv Yangmai 12 by using transgene-specific primers. P, Transformation vector pA25-TaPIE1 (positive control). The arrow indicates the transgene-specific amplification band (296 bp). B, Southern-blot assay of the OE and wild-type plants. Twenty micrograms of genomic DNA digested by the restriction enzyme SpeI was hybridized with the probe derived from the TaPIE1 transgene-specific fragment (296 bp). C, RT-PCR and qRT-PCR analyses of TaPIE1 transcription in the OE and wild-type plants. The relative transcript abundance of TaPIE1 in the three OE transgenic lines was compared with that in wild-type wheat (set to 1). D, Chlorotic mosaic symptoms on the fourth leaves of BSMV:00-infected (control) and BSMV:TaPIE1-infected (TaPIE1-underexpressing [UE]) wheat cv Yangmai 12 seedlings. The photographs were taken at 14 dpi. E and F, RT-PCR and qRT-PCR analyses of relative transcript levels of the BSMV COAT PROTEIN (CP) gene (E) and TaPIE1 (F) in the fourth leaves of the UE and control seedlings. The relative transcript abundance of TaPIE1 in the UE plants was related to that in a control plant (set to 1). The amplification of wheat ACTIN (TaActin) was used as an internal control to normalize all the data. Significant differences between the OE and wild-type plants or the UE and control plants were derived from the results of three independent replications (Student’s t test: **P < 0.01). Error bars indicate se. [See online article for color version of this figure.]