Abstract
Thiazide diuretics and statins are used to improve cardiovascular outcomes, but may also cause type 2 diabetes (T2DM), although mechanisms are unknown. Gene expression studies may facilitate understanding of these associations. Participants from ongoing population-based studies were sampled for these longitudinal studies of peripheral blood microarray gene expression, and followed to incident diabetes. All sampled subjects were statin or thiazide users. Those who developed diabetes during follow-up comprised cases (44 thiazide users; 19 statin users), and were matched to drug-using controls who did not develop diabetes on several factors. Supervised normalization, surrogate variable analyses removed technical bias and confounding. Differentially-expressed genes were those with a false discovery rate Q-value<0.05. Among thiazide users, diabetes cases had significantly different expression of CCL14 (down-regulated 6%, Q-value=0.0257), compared with controls. Among statin users, diabetes cases had marginal but insignificantly different expression of ZNF532 (up-regulated 15%, Q-value=0.0584), CXORF21 (up-regulated 11%, Q-value=0.0584), and ZNHIT3 (up-regulated 19%, Q-value=0.0959), compared with controls. These genes comprise potential targets for future expression or mechanistic research on medication-related diabetes development.
Keywords: Type 2 diabetes, statins, thiazide diuretics, whole blood, gene expression, microarray, supervised normalization, surrogate variable analysis, chemokine ligand 14, zinc finger proteins
Introduction
Thiazide diuretics and statins are used to treat hypertension or hyperlipidemia to reduce risk of cardiovascular disease (CVD). However, observational studies and clinical trials have also reported adverse consequences of these two drugs, including development of hyperglycemia, insulin resistance, and type 2 diabetes mellitus (T2DM), all of which may increase CVD risk [1-3]. Characterizing the way in which use of statins and thiazide diuretics affect development of disease states such as T2DM may improve understanding of the systemic effects these medications have on patients. To date, mechanistic pathways for these associations are not well understood; in particular, gene expression differences related to use of these medications and incident T2DM are largely unknown.
Relative gene expression levels are a biomarker representing the downstream confluence of genetic and environmental exposures, and as such may be used to characterize underlying molecular processes through which environment—such as use of medications—influences development of disease—such as T2DM [4]. We examined global gene expression in peripheral whole blood samples in an attempt to identify gene expression effects characterizing the way in which thiazide diuretics and statins may influence development of incident T2DM.
Materials/methods
Study participants were sampled from ongoing, population-based, case-control studies of cardiovascular diseases that constitute the Heart and Vascular Health Study (HVH). HVH participants are members of Group Health Cooperative (GHC) in western Washington State who were 21-79 years of age at study recruitment. HVH study participants were either diagnosed with an incident cardiovascular event (myocardial infarction, stroke, venous thrombosis, or atrial fibrillation) between 1995 and 2009 or were sampled at random from GHC members who had no history of these cardiovascular events. HVH subjects with and without cardiovascular events were frequency matched by age (within decade), sex, and year of identification. HVH participants were eligible for the current gene expression studies if they provided blood samples for RNA extraction. All participants provided written, informed consent to participate in genetic studies and the GHC IRB approved study protocols.
Participants in the current, longitudinal gene expression studies were current users of either thiazide diuretics or statins and were free of type 2 diabetes (T2DM) at the time of their blood draw, which was set as the current study baseline. “Current” use of medication was defined at blood draw date using GHC computerized pharmacy records if (1) the patient had at least two prescriptions filled before the blood draw date, and (2) he/she received enough medications in the last prescription filled to last until the blood draw date, assuming 80% compliance with prescribing instructions. Diabetes-negative status was defined based on no self-report of diabetes, no prior diagnoses in their medical record, and never having taken diabetes medications as of their blood draw date. Participants who were taking oral anticoagulants, glucocorticoids, methotrexate, purine antimetabolites, or oral contraceptives at the time of blood draw were excluded.
T2DM cases were participants who were diagnosed with incident T2DM after their blood draw date, and were identified based on a first-time use of insulin, metformin, glyburide, and/or glipizide. Controls were individuals who did not develop T2DM after their blood draw date, and were matched to T2DM cases based on time taking the exposure medications before blood draw date (lead time), follow-up time after blood draw date, blood draw year, race, baseline body mass index (BMI), and history of a cardiovascular event. Thiazide diuretic users who qualified as cases (N=44) were sampled and matched with thiazide diuretic-using controls directly from the HVH study population (N=44). Statin users who qualified as cases (N=19) and controls (N=37) were sampled from ongoing gene expression studies, among HVH participants. The other ongoing gene expression studies from which the current statin using group was selected included case-control studies for myocardial infarction, stroke, atrial fibrillation, and venous thromboembolism, as well as a cohort study on estrogen use. Because of the subject selection process, there was some overlap (N=25) between the two subject populations.
Peripheral whole blood samples were collected using PaxGene tubes (Qiagen Inc; Valencia, CA) between 2003 and 2012. RNA was extracted using Qiagen kits, per lab protocol at the Fred Hutchinson Cancer Research Center (FHCRC; Seattle, WA). RNA quality assessment and gene expression profiling (using microarray and qRT-PCR experiments) were conducted at the Biochemistry and Expression Laboratories (Southern California). RNA sample quality was assessed using Agilent BioAnalyzer (Agilent Technologies; Santa Clara, CA) and RNA Integrity Numbers (RINs). All samples were shown to be free of extensive degradation. RNA samples were then amplified and reverse-transcribed using Illumina kits, per manufacturer instructions (Illumina Inc; San Diego, CA). Labeled cDNA were hybridized to Illumina Human HT-12 Version 3 & Version 4 Expression Beadchips. Spike-in oligonucleotide controls and technical replicates were used to ensure the absence of technical error. Array chip images were captured using an Illumina Beadarray scanner and control plots were visually inspected to confirm low background noise and high probe-binding specificity. Scanned microarray chip images were processed using Illumina GenomeStudio to create expression data matrices. Signal intensities of missing probes were imputed by Genome Studio using the kNN algorithm. Gene-level expression matrices were calculated without background correction from probe-level expression matrices using the average of intensity values for all measured and imputed probes for a given gene. The final gene set used in statistical analyses included those that surpassed QC minima for both chip versions; thiazide diuretic analyses (21,601 genes) and statin analyses (18,738 genes).
Log-transformed gene-level expression data matrices were pre-processed and analyzed using Bioconductor packages in the R statistical computing environment (R Foundation for Statistical Computing, Vienna, Austria), including supervised normalization of microarrays (SNM), surrogate variable analysis (SVA), and Illumina microarray processing (lumi) packages [5-11].
SNM was used to account for potential sources of confounding as well as correction for multiple testing (based on approximately 20,000 tests) [12]. Besides matching factors (BMI, race, blood draw year, lead time, and follow-up time), a priori considered confounders included age, sex, other anti-hypertensive medication use, and batch (microarray chip). The thiazide diuretics analyses were not adjusted for statin use because it was not an a priori confounder; the statin analyses were adjusted for diuretics use because there was overlap in the subject populations, as the sampling for the statin analyses was done from among ongoing gene expression studies.
All primary analyses were based on regression models of expression intensity levels fit independently for each gene or RNA transcript, with intensity serving as the dependent variable and T2DM case/control status as the independent variable. Model parameters were estimated within the context of SNM, which includes adjustment for the matching factors and confounders listed previously, as well as adjustment for intensity-dependent effects [11]. SVA empirically assessed the residual data matrices for presence of surrogate variables, which could account for some of the technical and batch-based variability to which microarrays are prone, and final models adjusted for surrogate variables in addition to the other adjustments [9,10]. Analyses among thiazide users were adjusted for age; sex; BMI; chip; prior AF, MI, VTE, or stroke; time taking thiazides; follow-up time; use of any other hypertension medication; 5 SVs. Analyses among statin users were adjusted for age; sex; BMI; chip version; use of diuretics; use of any other hypertensive medications; 3 SVs. The SNM package used false discovery rate (FDR) methods to calculate Q-values for each gene based on ANOVA comparing nested models with and without the primary variable of interest, after the residual variability associated with adjustments was removed from the expression matrix [11]. Significant differential expression results for the current study were considered to be those with an estimated Q-value<0.05.
Results
Overall, study participants tended to be older adults, overweight, and users of preventative cardiovascular medications (Table 1). Cases (participants who developed T2DM) and controls were comparable to each other, with few exceptions. Among thiazide diuretic users, cases were younger than controls and the proportion using calcium-channel blockers was higher among cases than among controls. Age distributions of T2DM cases and controls, among statin-users, were similar; controls were more likely to be males than were cases. In addition, among statin users, T2DM cases were more likely to use diuretics than were controls.
Table 1.
Participant Characteristics
| Thiazide diuretic users, sampled from HVH population: | ||
|
| ||
| No T2D | Incident T2D | |
|
|
||
| N=44 | N=44 | |
|
| ||
| Male sex, n (%) | 19 (43) | 22 (50) |
| Age, mean (SD) | 71.9 (9.9) | 67.8 (8.3) |
| Prior AF, MI, VTE, stroke: n (%)‡ | 13 (30) | 13 (30) |
| BMI, mean (SD)‡ | 31.5 (4.1) | 31.5 (3.4) |
| Users of: | ||
| ACE-inhibitor, n (%) | 15 (34) | 13 (30) |
| Ca-channel blocker, n (%) | 3 (7) | 11 (25) |
| Beta blocker, n (%) | 19 (43) | 27 (61) |
| Statin, n (%) | 17 (39) | 23 (52) |
|
| ||
|---|---|---|
| Statin users, sampled from ongoing gene expression studies: | ||
|
| ||
| No T2 | Incident T2D | |
|
|
||
| N=37 | N=19 | |
|
| ||
| Male sex, n (%) | 27 (73) | 9 (47) |
| Age, mean (SD) | 71.4 (8.3) | 68.2 (6.5) |
| Prior AF, MI, VTE, stroke: n (%) | 0 | 0 |
| BMI, mean (SD) | 30.7 (5.6) | 32.4 (4.8) |
| Users of: | ||
| ACE-inhibitor, n (%) | 13 (35) | 9 (47) |
| Ca-channel blocker, n (%) | 6 (16) | 5 (26) |
| Beta blocker, n (%) | 20 (54) | 9 (47) |
| Diuretic, n (%) | 20 (54) | 16 (84) |
Matching factors during subject selection.
In QC analyses, using SNM-SVA packages, normalized array box plots, intensity-dependent effects per array, residual latent structure based on estimated principal components, convergence of the estimates of the proportion of non-differential or ‘null’ probes (π0), and the P-value distribution for the set of ‘null’ probes included in the estimation of π0, indicated that residual variability from non-biological sources was adequately removed from the comparison matrices (data not shown) [4-10].
The analyses among thiazide diuretic users (N=88), comparing incident T2DM cases (N=44) with controls (N=44), included microarray-based gene expression evaluation of 21,601 genes and RNA transcripts. The final estimated proportion of ‘null’ probes (π0) was 0.97. The QQ and volcano plots (Figure 1A, 1B) of these differential gene expression analyses also indicated that most gene expression comparisons were not statistically significant. Expression of one gene, CCL14 (H. sapiens chemokine C-C motif lig-and 14), was significantly lower among T2DM cases, after correction for multiple comparisons (P-value=1.22e-06; Q-value: 0.0257), although the actual fold-change was small (-6%) (Table 2). The other top ‘gene’, ranked by statistical significance, was MIR1226 (P-value=3.45e-05; Q-value: 0.3629) and was up-regulated among T2DM cases, compared with controls. However, this gene and others did not reach statistical significance after correction for multiple testing. The top gene, ranked by fold-change (but which was not statistically significant) was HLA-A29, highlighted on the volcano plot in blue (Figure 1B).
Figure 1.
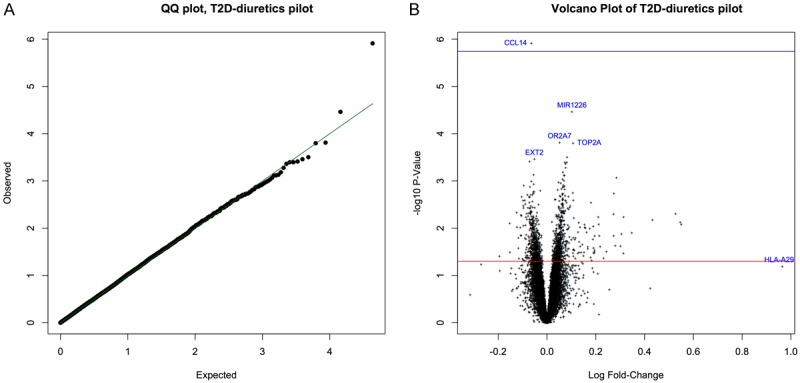
A: Quantile-quantile (QQ) plot of expected and observed P-values for each gene in the thiazide diuretics study comparing incident T2DM cases with non-cases. B: Volcano plot of (-log, base 10) P-value versus (log) fold-change coparing incident T2DM cases with non-cases in the thiazide diuretics study. Top 5 genes (based on statistical test) and single top gene (based on fold-change) highlighted in blue.
Table 2.
Top 10 Genes, Comparing T2DM Cases with Controls
| Thiazide diuretic Study† | ||||
|
| ||||
| Gene | Ratio of Means | % FC | P-value, unadjusted | FDR/Q-value |
| CCL14 | 0.938 | -6% | 1.22E-6 | 0.026 |
| MIR1226 | 1.108 | 11% | 3.45E-5 | 0.363 |
| OR2A7 | 1.053 | 5% | 0.0002 | 0.837 |
| TOP2A | 1.113 | 11% | 0.0002 | 0.837 |
| EXT2 | 0.949 | -5% | 0.0003 | 0.870 |
|
| ||||
|---|---|---|---|---|
| Statin Study‡ | ||||
|
| ||||
| Gene | Fold change | % FC | P-value, unadjusted | FDR/Q-value |
|
| ||||
| ZNF532 | 1.150 | 15% | 4.47E-6 | 0.058 |
| CXORF21 | 1.105 | 11% | 7.02E-6 | 0.058 |
| ZNHIT3 | 1.185 | 19% | 1.73E-5 | 0.096 |
| EBAG9 | 1.098 | 10% | 9.34E-5 | 0.332 |
| PHF10 | 0.886 | -11% | 0.0001 | 0.332 |
Analyses among thiazide users: adjusted for age; sex; BMI; chip; prior AF, MI, VTE, stroke; time taking thiazides; follow-up time; use of any other hypertension medication; 5 SVs.
Analyses among statin users: adjusted for age; sex; BMI; chip version; use of diuretics; use of any other hypertensive medications; 3 SVs.
The analyses among statin users (N=56), comparing incident T2DM cases (N=19) with controls (N=37), included microarray-based RNA expression evaluation for 18,738 genes and RNA transcripts. The final estimated proportion of ‘null’ genes (π0) was 0.89. The QQ and volcano plots (Figure 2A, 2B) of these differential expression analyses also indicated that most gene expression comparisons were not statistically significant. No gene expression difference between cases and controls surpassed statistical significance after multiple testing corrections. However, some genes had marginally significant Q-values. These included ZNF532 (zinc finger protein 532; up-regulated, Q-value=0.0584), CXORF21 (chromosome X open reading frame 21; up-regulated, Q-value=0.0584), and ZNHIT3 (zinc finger, HIT-type containing 3; up-regulated, Q-value=0.0959).
Figure 2.
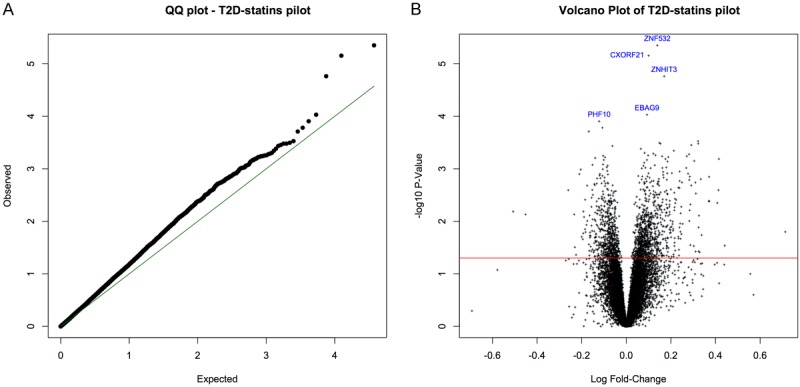
A: Quantile-quantile (QQ) plot of expected and observed P-values for each gene in the statins study comparing incident T2DM cases with non-cases. B: Volcano plot of (-log, base 10) P-value versus (log) fold-change coparing incident T2DM cases with non-cases in the statins study. Top 5 genes (based on statistical test) highlighted in blue.
Table 3 provides detailed information for each of the top 5 genes for both the thiazide diuretics study and the statins study.
Table 3.
Detailed Information for Top 10 Gene Results
| Gene | Illumina Search Key | Chr. | Gene Full Name+ | Aliases |
|---|---|---|---|---|
| CCL14 | NM_032962.2 | H. sapiens chemokine (C-C motif) ligand 14, transcript variant 2 | SCYL2; NCC-2; MCIF; HCC-3; CC-3; SCYA14; CC-1; SY14; NCC2; HCC-1; CKb1 | |
| MIR1226 | NR_031595.1 | H. sapiens microRNA 1226, miRNA | ||
| OR2A7 | NM_001005328.1 | 7 | H. sapiens olfactory receptor, family 2, subfamily A, member 7 | HSDJ0798C17; OR2A21 |
| TOP2A | NM_001067.2 | 17 | H. sapiens topoisomerase (DNA) II alpha | TOP2; TP2A |
| EXT2 | NM_000401.2 | 11 | H. sapiens exostoses (multiple) 2, transcript variant 1 | SOTV |
| ZNF532 | NM_018181.4 | 18 | H. sapiens zinc finger protein 532 | FLJ10697 |
| CXORF21 | NM_025159.1 | X | H. sapiens chromosome X open reading frame 21 | FLJ11577 |
| ZNHIT3 | NM_001033577.1 | 17 | H. sapiens zinc finger, HIT type 3, transcript variant 1 | TRIP3 |
| EBAG9 | NM_198120.1 | 8 | H. sapiens estrogen receptor binding site associated, antigen, 9, transcript variant 2 | RCAS1; EB9; PDAF |
| PHF10 | NM_133325.1 | 6 | H. sapiens PHD finger protein 10, transcript variant 2 | XAP135; FLJ10975; MGC111009 |
All messenger RNA (mRNA) except where noted as micro RNA (miRNA).
Discussion
Findings from our study suggest that, among users of thiazide diuretics or statins, there may be significant differences in gene expression in peripheral blood of individuals who subsequently developed incident diabetes (T2DM), compared with those who did not. The CCL14 gene was differentially expressed (down-regulated) among thiazide diuretic users who developed T2DM, while some evidence for differential expression of ZNF532 (up-regulated), CXORF21 (up-regulated) and ZNHIT2 (up-regulated) was observed among statin users who developed T2DM, compared to their respective controls. Identified genes are novel in terms of their associations with T2DM, and need to be confirmed in additional investigations.
Previous studies have reported increased risk of incident T2DM ranging from 20-80% in relation to use of thiazide diuretics [13-18]. In one major trial, thiazide use was associated with 43% greater risk over 4 years, compared with use of another antihypertensive, but, overall, comparison groups vary widely and methodological issues limit the conclusiveness of the published data [1,3,19]. Hypokalemia and metabolic alkalosis resulting from thiazide diuretic use may lead to glucose intolerance and hyperglycemia [20]. Previously suggested mechanisms for these associations include inflammation, activation of the renin-angiotensin-aldosterone system, and decreases in adiponectin [20,21]. The CCL14 gene, differentially expressed in our study, is one in a cluster of cytokine genes that is expressed in several tissues (such as spleen, liver, skeletal and heart muscle, gut, bone marrow), and its functions involve calcium ion homeostasis, inflammation/immune response, and cell signaling [22]. While previous studies reported associations of CCL14 genetic variation and CCL14 protein levels with human diseases, such as bronchiolitis and SLE, to our knowledge it has not previously been associated with T2DM [23,24]. Given that inflammation is one of the hypothesized mechanisms linking thiazide diuretics with T2DM, and that previous reports have found inflammation and release of cytokines/chemokines to be involved in T2DM pathogenesis, further research that explores the role of CCL14 in T2DM pathogenesis and the relationships between thiazide diuretics and T2DM is warranted [25].
The risk of incident T2DM associated with statin use has been estimated to range from 9-70%; put another way, treatment of an estimated 255 patients with statins for 4 years may result in 1 additional case of T2DM [2,26,27]. Statins may lead to reduced insulin secretion through competitive binding of HMG-CoA reductase or cytotoxicity, both of which may inhibit the normal function of pancreatic cells that regulate calcium channels and insulin secretion [28]. Statins may also disrupt insulin sensitivity by blocking insulin receptors via disruption of the Pi3K-pAkt-mTOR pathway [29]. In our study, expression of three genes, ZNF532, CXORF21, and ZNHIT3 was marginally associated with incident T2DM among statin users. Little is known about ZNF532 and CXORF21 apart from their potential roles in transcription regulation [30,31]. ZNHIT3, another transcription regulator, interacts with well-described transcription factors (such as THRB, HNF4A, and TRIP3) which have roles in lipid and glucose metabolism [32-34]. None of the three genes that we identified, however, has previously been directly linked with glucose metabolism, insulin resistance, or reduced insulin secretion. Further studies are needed to evaluate their relevance to incident T2DM.
Drug-specific effects of statins on insulin sensitivity, and thus T2DM risk, have been previously suggested [22,24-26]. For instance, pravastatin may improve insulin sensitivity while simvastatin, rosuvastatin, and atorvastatin may worsen it [26,35-37]. We did not evaluate these drug-specific effects in relation to gene expression differences because the current study was conducted among members of a single health plan with an established formulary, and so there was very limited variability in the different statins (atorvastatin, lovastatin, pravastatin) that were used.
Some limitations of our study warrant mention. It is possible that there are gene expression differences between cases and controls that did not result from use of statins or thiazides; future research may examine prospective, longitudinal data to confirm these findings. Case identification methods for these gene expression studies may limit external generalizability, especially for the statin analyses which were conducted from among subjects participating in ongoing gene expression studies. Pharmacy record data were used to identify T2DM medication use as a proxy for disease status, which may misclassify preclinical, undiagnosed, or non-compliant diabetes patients. A priori hypothesized confounders were evaluated, but unmeasured confounders may exist. Differential blood cell populations may exist between T2DM cases and controls, but it is not clear whether such a difference would represent a confounder or part of the causal pathway between medication use and incident diabetes; there was not statistical power to examine this question. It is possible that there is effect modification by the length of time having taken medications before the blood draw date (lead time), but it was not possible to investigate this question in the current study due to lack of statistical power. Although peripheral whole blood is likely to be reflective of systemic effects on gene pathways that are important in T2DM risk, it may not fully or adequately represent organ-level gene expression differences important to development of T2DM.
The high prevalence of thiazide diuretic and statin use underlines the need to develop a better understanding of adverse consequences of these medications. Our study and its findings may contribute to these efforts. Future research in gene expression has great potential to further provide insight on these relationships between thiazide diuretics or statins and T2DM, and to contribute to understanding of diabetes pathology, pharmacoepidemiology, and genetic epidemiology.
Acknowledgements
A. Suchy-Dicey conducted this work while supported by the NHLBI Cardiovascular Disease Training Grant, NIH I-T32-HL07902. She received the 2012-2013 Magnuson Scholarship Award from the University of Washington. This study was supported by grants from the National Health Lung and Blood Institute (HL73410, HL60739, HL68639, HL74745, and HL68986). The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Disclosure of conflict of interest
Bruce Psaty served on the Data Safety Monitoring Board for a clinical trial of a device funded by the manufacturer (2011 LifeCor). There are no other potential conflicts to disclose.
References
- 1.Barzilay JI, Davis BR, Cutler JA, Pressel SL, Whelton PK, Basile J, Margolis KL, Ong ST, Sadler LS, Summerson J ALLHAT Collaborative Research Group. Fasting glucose levels and incident diabetes mellitus in older nondiabetic adults randomized to receive 3 different classes of antihypertensive treatment: a report from the Antihypertensive and Lipid-Lowering Treatment to Prevent Heart Attack Trial (ALLHAT) Arch Intern Med. 2006;166:2191–201. doi: 10.1001/archinte.166.20.2191. [DOI] [PubMed] [Google Scholar]
- 2.Sattar N, Preiss D, Murray HM, Welsh P, Buckley BM, de Craen AJ, Seshasai SR, McMurray JJ, Freeman DJ, Jukema JW, Macfarlane PW, Packard CJ, Stott DJ, Westendorp RG, Shepherd J, Davis BR, Pressel SL, Marchioli R, Marfisi RM, Maggioni AP, Tavazzi L, Tognoni G, Kjekshus J, Pedersen TR, Cook TJ, Gotto AM, Clearfield MB, Downs JR, Nakamura H, Ohashi Y, Mizuno K, Ray KK, Ford I. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet. 2010;375:735–42. doi: 10.1016/S0140-6736(09)61965-6. [DOI] [PubMed] [Google Scholar]
- 3.Padwal R, Laupacis A. Antihypertensive therapy and incidence of type 2 diabetes: a systematic review. Diabetes Care. 2004;27:247–55. doi: 10.2337/diacare.27.1.247. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh S, Dent R, Harper ME, Gorman SA, Stuart JS, McPherson R. Gene expression profiling in whole blood identifies distinct biological pathways associated with obesity. BMC Med Genomics. 2010;3:56. doi: 10.1186/1755-8794-3-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 7.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentleman RHK. R 1.5 and the Bioconductor 1.0 releases. Comput Stat Data An. 2002;39:558–9. [Google Scholar]
- 9.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genetics. 2007;3:1724–35. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mecham BH, Nelson PS, Storey JD. Supervised normalization of microarrays. Bioinformatics. 2010;26:1308–15. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lunceford JK, Chen G, Hu PH, Mehrotra DV. Evaluating surrogate variables for improving microarray multiple testing inference. Pharm Stat. 2011;10:302–10. doi: 10.1002/pst.466. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee R, Yeh HC, Shafi T, Selvin E, Anderson C, Pankow JS, Miller E, Brancati F. Serum and dietary potassium and risk of incident type 2 diabetes mellitus: The Atherosclerosis Risk in Communities (ARIC) study. Arch Intern Med. 2010;170:1745–51. doi: 10.1001/archinternmed.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper-DeHoff RM, Wen S, Beitelshees AL, Zineh I, Gums JG, Turner ST, Gong Y, Hall K, Parekh V, Chapman AB, Boerwinkle E, Johnson JA. Impact of abdominal obesity on incidence of adverse metabolic effects associated with antihypertensive medications. Hypertension. 2010;55:61–8. doi: 10.1161/HYPERTENSIONAHA.109.139592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott WJ, Meyer PM. Incident diabetes in clinical trials of antihypertensive drugs: a network meta-analysis. Lancet. 2007;369:201–7. doi: 10.1016/S0140-6736(07)60108-1. [DOI] [PubMed] [Google Scholar]
- 16.Feero WG, Guttmacher AE, Collins FS. Genomic medicine--an updated primer. N Engl J Med. 2010;362:2001–11. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 17.Lam SK, Owen A. Incident diabetes in clinical trials of antihypertensive drugs. Lancet. 2007;369:1513–4. doi: 10.1016/S0140-6736(07)60697-7. author reply 4-5. [DOI] [PubMed] [Google Scholar]
- 18.Zillich AJ, Garg J, Basu S, Bakris GL, Carter BL. Thiazide diuretics, potassium, and the development of diabetes: a quantitative review. Hypertension. 2006;48:219–24. doi: 10.1161/01.HYP.0000231552.10054.aa. [DOI] [PubMed] [Google Scholar]
- 19.Stump CS, Hamilton MT, Sowers JR. Effect of antihypertensive agents on the development of type 2 diabetes mellitus. Mayo Clin Proc. 2006;81:796–806. doi: 10.4065/81.6.796. [DOI] [PubMed] [Google Scholar]
- 20.Manrique C, Johnson M, Sowers JR. Thiazide diuretics alone or with beta-blockers impair glucose metabolism in hypertensive patients with abdominal obesity. Hypertension. 2010;55:15–7. doi: 10.1161/HYPERTENSIONAHA.109.142620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bozkurt O, de Boer A, Grobbee DE, de Leeuw PW, Kroon AA, Schiffers P, Klungel OH. Variation in Renin-Angiotensin system and salt-sensitivity genes and the risk of diabetes mellitus associated with the use of thiazide diuretics. Am J Hypertens. 2009;22:545–51. doi: 10.1038/ajh.2009.38. [DOI] [PubMed] [Google Scholar]
- 22.Moelants EA, Mortier A, Van Damme J, Proost P. In vivo regulation of chemokine activity by post-translational modification. Immunol Cell Biol. 2013;91:402–7. doi: 10.1038/icb.2013.16. [DOI] [PubMed] [Google Scholar]
- 23.Siezen CL, Bont L, Hodemaekers HM, Ermers MJ, Doornbos G, Van’t Slot R, Wijmenga C, Houwelingen HC, Kimpen JL, Kimman TG, Hoebee B, Janssen R. Genetic susceptibility to respiratory syncytial virus bronchiolitis in preterm children is associated with airway remodeling genes and innate immune genes. Pediatr Infect Dis J. 2009;28:333–5. doi: 10.1097/INF.0b013e31818e2aa9. [DOI] [PubMed] [Google Scholar]
- 24.Vyshkina T, Sylvester A, Sadiq S, Bonilla E, Perl A, Kalman B. CCL genes in multiple sclerosis and systemic lupus erythematosus. J Neuroimmunol. 2008;200:145–52. doi: 10.1016/j.jneuroim.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saku K, Harada R, Yamamoto K, Ying H, Ozaki I, Arakawa K. Apolipoprotein AI mRNA levels in WHHL rabbits. Atherosclerosis. 1990;84:73–4. doi: 10.1016/0021-9150(90)90010-g. [DOI] [PubMed] [Google Scholar]
- 26.Mann D, Reynolds K, Smith D, Muntner P. Trends in statin use and low-density lipoprotein cholesterol levels among US adults: impact of the 2001 National Cholesterol Education Program guidelines. Ann Pharmacother. 2008;42:1208–15. doi: 10.1345/aph.1L181. [DOI] [PubMed] [Google Scholar]
- 27.Mascitelli L, Pezzetta F, Goldstein MR. Statins and risk of incident diabetes. Lancet. 2010;375:2140–1. doi: 10.1016/S0140-6736(10)60990-7. author reply 1-2. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa M, Okajima F, Inoue N, Motomura K, Kato T, Takahashi A, Oikawa S, Yamada N, Shimano H. Distinct effects of pravastatin, atorvastatin, and simvastatin on insulin secretion from a beta-cell line, MIN6 cells. J Atheroscler Thromb. 2006;13:329–35. doi: 10.5551/jat.13.329. [DOI] [PubMed] [Google Scholar]
- 29.Miraglia E, Hogberg J, Stenius U. Statins exhibit anticancer effects through modifications of the pAkt signaling pathway. Int J Oncol. 2012;40:867–75. doi: 10.3892/ijo.2011.1223. [DOI] [PubMed] [Google Scholar]
- 30.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T, Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai K, Isogai T, Sugano S. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. A data set of human endogenous protein ubiquitination sites. Mol Cell Proteomics. 2011 May;10:M110.002089. doi: 10.1074/mcp.M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwahashi H, Yamagata K, Yoshiuchi I, Terasaki J, Yang Q, Fukui K, Ihara A, Zhu Q, Asakura T, Cao Y, Imagawa A, Namba M, Hanafusa T, Miyagawa J, Matsuzawa Y. Thyroid hormone receptor interacting protein 3 (trip3) is a novel coactivator of hepatocyte nuclear factor-4alpha. Diabetes. 2002;51:910–4. doi: 10.2337/diabetes.51.4.910. [DOI] [PubMed] [Google Scholar]
- 33.Koppen A, Houtman R, Pijnenburg D, Jeninga EH, Ruijtenbeek R, Kalkhoven E. Nuclear receptor coregulator interaction profiling identifies TRIP3 as a novel peroxisome proliferator-activated receptor gamma cofactor. Mol Cell Proteomics. 2009 Oct;8:2212–26. doi: 10.1074/mcp.M900209-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–54. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 35.Baker WL, Talati R, White CM, Coleman CI. Differing effect of statins on insulin sensitivity in non-diabetics: a systematic review and meta-analysis. Diabetes Res Clin Pract. 2010;87:98–107. doi: 10.1016/j.diabres.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Culver AL, Ockene IS, Balasubramanian R, Olendzki BC, Sepavich DM, Wactawski-Wende J, Manson JE, Qiao Y, Liu S, Merriam PA, Rahilly-Tierny C, Thomas F, Berger JS, Ockene JK, Curb JD, Ma Y. Statin use and risk of diabetes mellitus in postmenopausal women in the Women’s Health Initiative. Arch Intern Med. 2012;172:144–52. doi: 10.1001/archinternmed.2011.625. [DOI] [PubMed] [Google Scholar]
- 37.Moutzouri E, Liberopoulos E, Mikhailidis DP, Kostapanos MS, Kei AA, Milionis H, Elisaf M. Comparison of the effects of simvastatin vs. rosuvastatin vs. simvastatin/ezetimibe on parameters of insulin resistance. Int J Clin Pract. 2011;65:1141–8. doi: 10.1111/j.1742-1241.2011.02779.x. [DOI] [PubMed] [Google Scholar]


