Abstract
The formation and characterization of an activated complex of the visual pigment rhodopsin and its downstream signaling partner transducin has been the subject of intense focus by several research groups. While the subunit composition of the activated complex is still the subject of some controversy, our laboratory [Xie, G., D’Antona, A. M., Edwards, P. C., Fransen, M., Standfuss, J., Schertler, G. F. X., and Oprian, D. D. (2011) Biochemistry 50, 10399–10407] and that of Ernst et al. [Ernst, O. P., Gramse, V., Kolbe, M., Hofmann, K. P., and Heck, M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 10859–10864] find that the two proteins are present in a 1/1 molar ratio. Unfortunately, these data could not distinguish a 1/1 complex from 2/2 or 3/3 and so on. For this reason, we reinvestigated the issue of stoichiometry in the activated complex, exploiting the ability of Nanodisc lipid bilayers to isolate single molecules of rhodopsin. We show here that the purified complex in Nanodiscs contains an activated rhodopsin with a covalently-bound all-trans-retinal chromophore; that transducin has an empty nucleotide-binding pocket; that the isolated complex is active and dissociates upon addition of guanine nucleotide; and finally that the stoichiometry corresponds to exactly one molecule of rhodopsin and one molecule of transducin.
The visual pigment rhodopsin of vertebrate rod photoreceptor cells is a prototypical member of the large class of G protein-coupled receptors(1). It is composed of an apoprotein, opsin, and an 11-cis-retinal chromophore covalently attached to a Lys residue (Lys296) in the 7th transmembrane (TM) segment of the protein by means of a protonated Schiff base linkage(2). The chromophore undergoes photoisomerization to the all-trans form triggering a conformational change in rhodopsin that leads to activation of the G protein transducin (Gt) and initiation of a signaling cascade that leads ultimately to closure of cGMP-gated channels in the rod outer segment, hyperpolarization of the plasma membrane, and inhibition of glutamate release from the synaptic terminus(3).
The critical interaction of Gt with activated rhodopsin, or metarhodopsin II (R*)(4), has been the subject of numerous studies, the most recent of which have focused on isolation and purification of the activated complex in detergent solution, and determination of its subunit composition. While the stoichiometry of rhodopsin and Gt in the activated complex remains a subject of some controversy(5–7), our laboratory and others report a molar ratio of 1/1 for rhodopsin and Gt(8, 9). Importantly, however, these data do not unambiguously determine the absolute subunit composition in the complex, as a 1/1 molar ratio cannot be distinguished from 2/2, 3/3, and so on. For this reason, we re-examined the question of subunit stoichiometry, this time exploiting the ability of Nanodiscs to isolate single molecules of rhodopsin(10–15).
We report here the purification and characterization of an activated complex of R* and Gt in Nanodiscs using native Gt and a constitutively active mutant of rhodopsin(16) containing also an engineered disulfide bond known to confer heightened thermal stability on the protein(17), as was previously described for purification of the complex in detergent solution(9). We show that the Nanodisc complex forms only if rhodopsin is in the activated state and that the nucleotide-binding site in Gt is empty. Most importantly, we show unambiguously that the complex is composed of 1 molecule of rhodopsin and 1 molecule of Gt.
EXPERIMENTAL PROCEDURES
Materials
The anti-rhodopsin monoclonal antibody 1D4(18, 19) was from the National Cell Culture Center (Minneapolis, MN). CNBr-activated Sepharose 4B was from GE Healthcare. The 1D4-Sepharose 4B used for purification of rhodopsin and the R*/Gt complex in Nanodiscs was prepared as previously described(20). 1D4-peptide, a synthetic octapeptide (ETSQVAPA) corresponding to the 1D4-epitope of the C-terminal eight amino acids of opsin, was used for elution of rhodopsin from the 1D4-Sepharose 4B matrix(20). Concanavalin A-Sepharose 4B (ConA-Sepharose) was a product of Sigma. 11-cis- and all-trans-retinal were synthesized and purified according to published procedures, as described by Xie et al. 2003(17). [α−32P]-GTP (3000 Ci/mmol) and [35S]-GTPγS (1250 Ci/mmol) were purchased from Perkin Elmer. GTP and GTPγS were from Amersham and Sigma, respectively. The membrane scaffolding protein, MSP1D1, was expressed and purified as described(21). 1-Palmitoyl-2-oleoylsn-glycero-3-phosphocholine (POPC) was from Avanti Polar Lipids, Inc. (Alabaster, AL). Other reagents were as reported earlier(9).
Expression and Purification of Rhodopsin
All experiments in this study were conducted with the N2C,E113Q,D282C triple mutant of rhodopsin. This mutant combines the constitutive activity of the E113Q mutant(16) with the increased thermal stability of the N2C,D282C mutant(17). The N2C,E113Q,D282C mutant was expressed transiently in HEK293S-GnT1− cells following transfection with calcium phosphate(22). The pigment was then purified from transfected cells using 1D4-immunoaffinity chromoatography essentially as described(9). Assembly and purification of the Rho/Gt complex on ConA-Sepharose began with N2C,E113Q,D282C mutant that had been previously purified from HEK293S-GnT1− cells using immunoaffinity chromatography on the 1D4-column.
Preparation of Rhodopsin-Embedded Nanodiscs
Rhodopsin-embedded Nanodiscs were prepared as described(11). While the interaction of rhodopsin and arrestin is known to be highly sensitive to lipid composition(12, 13), the interaction of rhodopsin and Gt is not, and for this reason we did not explore the effect of different lipids on formation of the R*/Gt complex. We chose POPC for continuity with previous work(11). POPC Nanodisc self-assembly was performed at ratios of 70/1/0.07 POPC/MSP1D1/rhodopsin to ensure that only one rhodopsin was present per Nanodisc. Rhodopsin, purified with 1D4-Sepharose, was first applied to a 40 kDa molecular weight cut-off (MWCO) centrifugal device (Amicon Ultra from Millipore) and washed with several volumes of 5 mM Hepes, pH 7.5, 0.1 mM MgCl2 (Buffer A) containing 0.02% (w/v) dodecyl β-D-maltosde (DDM) to remove the 1D4-peptide. The protein was then mixed with MSP1D1 and POPC (Avanti Polar Lipids) and incubated for 30 min on ice. An equal volume of prewashed Biobeads SM-2 (Bio-Rad) was then added and incubated overnight at 4°C to remove detergent. Biobeads were separated by brief centrifugation and the supernatant fraction containing rhodopsin Nanodiscs was collected. The final concentration of rhodopsin in the Nanodisc assembly mixture was typically about 7 µM. Stock solutions of 0.1 M POPC were prepared in 20 mM Hepes, pH 7.5, 0.1 mM MgCl2, and 120 mM NaCl (Buffer B) containing 0.2 M CHAPS.
Purification of Native Gt
Gt was purified from frozen bovine retinae (Schenk Packing Co.) according to previously published procedures(23). Rod photoreceptor cell outer segment (ROS) membranes were exposed to light, washed to remove associated proteins, and then treated with 40 µM GTP in 10 mM Tris, pH 7.4, 0.1 mM EDTA, and 2 mM dithiothreitol (DTT) to elute Gt. ROS were then removed by centrifugation. The supernatant fraction was concentrated with a 50-kDa MWCO centrifugal device, filtered through a 0.22 µm membrane filter (Steriflip from Millipore), dialyzed against 5 mM Tris, pH 7.4, 1 mM MgCl2, 0.5 mM DTT plus 50% (v/v) glycerol, and then stored at −20°C until use. The concentration of Gt was determined by absorption spectroscopy (ε280 for GtGDP = 93,570 M −1 cm−1), Lowry assay(24), and active-site titration with radiolabeled [35S]-GTPγS of known specific activity. Concentrations determined by Lowry assay or absorption spectroscopy were typically within 5 to 10% of the value as determined by active-site titration. The same procedure was followed for the preparation of Gt containing [α-32P]-GDP in the nucleotide binding site except that 40 µM [α32P]-GTP (0.31 Ci/mmol) was used instead of nonradiolabeled GTP for the release of Gt from ROS membranes.
Absorption Spectroscopy
UV-visible absorption spectra were recorded using a Hitiachi model U-3210 that was specifically modified by the manufacturer for use in a dark room. Data were collected with a microcomputer and evaluated with Graphpad Prism software (Graphpad Software, San Deigo, CA). All samples were recorded at 25°C with a path length of 1 cm. The molar absorption coefficient for the rhodopsin triple mutant in Nanodiscs was determined by acid trapping of the chromophore as described(25) except that 50 mM sodium phosphate buffer, pH 3.5, containing 0.5% (w/v) SDS was used. Molar absorption coefficients for MSP1D1 (ε280 = 21,430 M−1·cm−1) and nucleotide-free Gt, (Gtempty; ε280 = 87,800 M−1 cm−1) were calculated with the ExPASy ProtParam tool(26) which uses the Edelhoch method(27) with extinction coefficients for Trp and Tyr determined by Pace et al.(28). GTP ε280 = 5,770 M−1 cm−1 was measured experimentally assuming ε253 = 13,700 M−1•cm−1 and was used to calculate ε280 for GtGDP = 93,570 M−1 cm−1.
Gt Activation Assay
The ability of rhodopsin to catalyze the light-dependent exchange of radiolabeled [35S]-GTPγS for bound GDP in Gt was monitored using a filter-binding assay as described(29). To assay the R*/Gt complex for activity, a similar procedure was used except that the purified complex was diluted to a final concentration of 30 to 40 nM rhodopsin in 10 mM Tris, pH 7.5, 40 µM [35S]-GTPγS (1.42 Ci/mmol), 100 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA and 1 mM DTT without additional Gt. Reactions were incubated for 5 min at 25°C before aliquots were withdrawn and applied to nitrocellulose filters.
Stoichiometry of the Activated Complex
The relative stoichiometry of rhodopsin, transducin, and MSP1D1 in the activated complex was determined by densitometry of protein bands following electrophoretic separation on SDS-PAGE gels. Purified complex was visualized by staining with Coomasie Blue. Band densities were quantified from background-subtracted digitized gel images using Quantity One software (Bio-Rad, Hercules, CA). Protein quantities from unknown samples were determined using a standard curve of known amounts of protein as determined by UV-visible absorption spectroscopy.
RESULTS
Preparation of Nanodiscs Containing the Rhodopsin N2C,E113Q,D282C Triple Mutant
Nanodiscs containing the rhodopsin N2C,E113Q,D282C triple mutant (hereafter referred to simply as “rhodopsin”) were prepared from a mixture of POPC/MSP1D1/rhodopsin in the ratio 70/1.0/0.07 to ensure that each Nanodisc contained only one molecule of rhodopsin, as has been described(11). Empty Nanodiscs were removed by purifying the rhodopsin-embedded Nanodiscs by chromatography on a 1D4-Sepharose column.
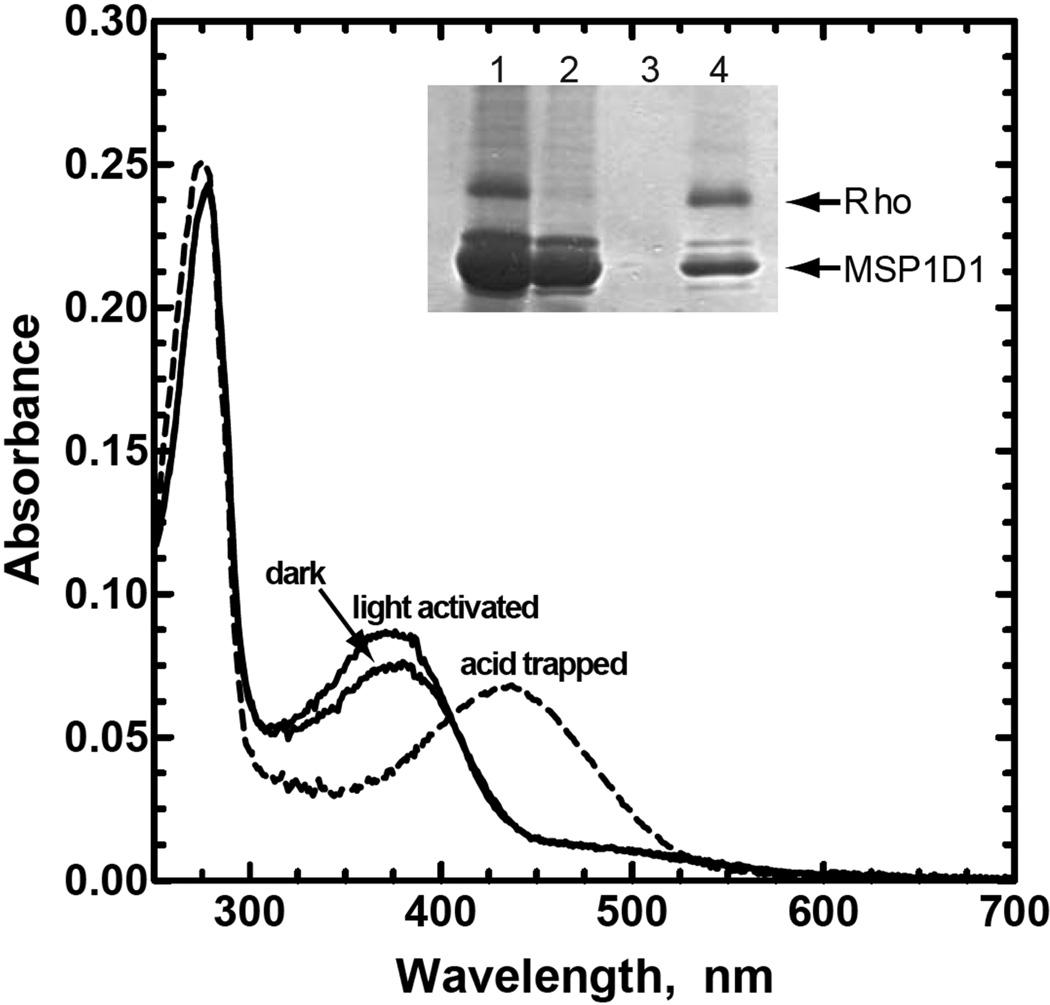
As is shown in Fig. 1, absorption spectra for rhodopsin in Nanodiscs reconstituted with 11-cis-retinal chromophore at pH 7.5 displays an absorption maximum (λmax) at 380 nm, characteristic of the unprotonated Schiff base form of the chromophore, as well as a second λmax at 280 nm characteristic of protein. The A280nm/A380nm ratio was larger for the protein in Nanodiscs as compared to a detergent-solubilized preparation in DDM micelles(9, 17) because of the contribution at 280 nm from the co-purified MSP1D1 (Fig. 1, dark spectrum). Subtracting the contribution of rhodopsin to the absorbance at 280 nm (assuming an A280 nm/A380 nm ratio of 1.7 for purified rhodopsin) provides an initial estimation of two moles of MSP1D1 per mole of rhodopsin, consistent with the Coomasie Blue-stained SDS-PAGE gel shown in the inset of Fig. 1, and with previously published results for Nanodiscs prepared under similar conditions(11).
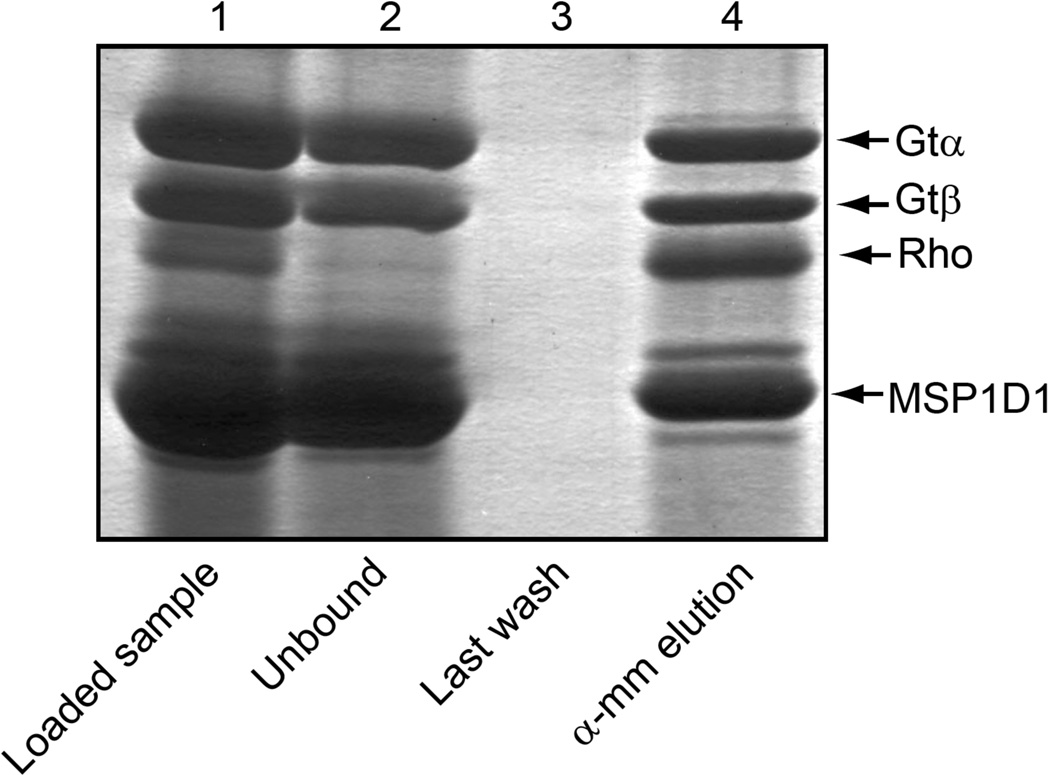
Figure 1.
Preparation of rhodopsin-containing Nanodiscs. The N2C,E113Q,D282C rhodopsin triple mutant was reconstituted with 11-cis-retinal, inserted into the lipid bilayer of Nanodiscs, and purified by immunoaffinity chromatography on 1D4-Sepharose. Dark, Absorption spectrum of the rhodopsin-containing Nanodiscs purified in the dark at pH 7.5; light activated, spectrum following 2 min illumination with light from a 300w tungsten bulb; acid trapped, spectrum immediately following the addition of 0.5% SDS and 50 mM sodium phosphate buffer, pH 3.5, to the light exposed sample. The acid trapped spectrum has been corrected for the effect of dilution. Inset, SDS-PAGE analysis of the steps used for preparation and purification of the rhodopsin containing Nanodiscs. Lane 1, rhodopsin-MSP1D1 mixture following detergent removal but before loading onto the 1D4-Sepharose column; lane 2, unbound material from the 1D4-Sepharose column; lane 3, last wash before elution; and lane 4, 1D4-peptide eluate containing purified N2C,E113Q,D282C rhodopsin triple mutant in MSP1D1 Nanodiscs. Protein bands in the gel were visualized with Coomassie Blue.
Illumination of the sample increased the absorbance at 380nm (light activated spectrum). Subsequent acid denaturation by addition of 0.5% SDS in 50mM sodium phosphate buffer at pH 3.5 converted the λmax from 380 nm to a new peak at about 430 nm (acid trapped spectrum), characteristic of a retinylidene protonated Schiff base attached to the denatured protein(25). The absorption coefficients at 380 nm for the dark-adapted and light-activated forms of rhodopsin in Nanodiscs were determined to be 35,200 M−1 cm−1 and 41,500 M−1 cm−1, respectively, using ε430 = 32,500 M−1 cm−1 for the chromophore under acid/denaturing conditions.
Activation of Gt in the Presence of the 1D4-Antibody
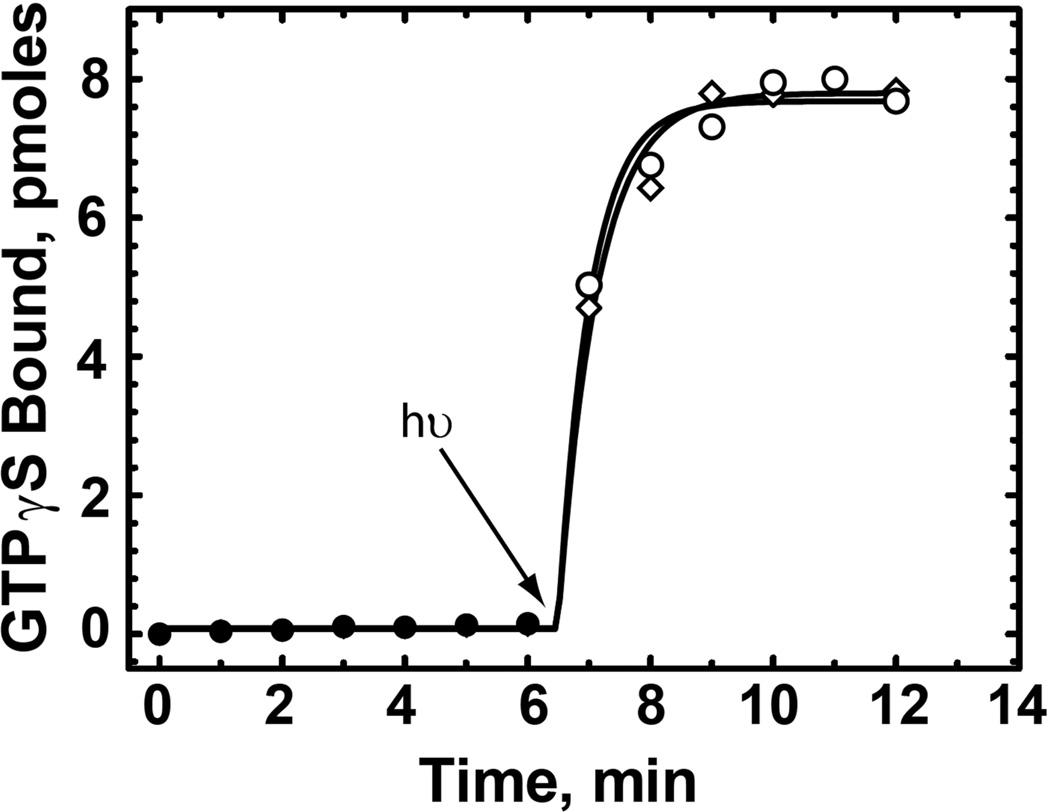
Our approach in isolation of the R*/Gt complex is to make use of the tight binding between R* and Gt in the absence of nucleotide to form the complex and then exploit the specific interaction between rhodopsin and the 1D4-antibody for purification. This strategy rests on the principle that the binding of the 1D4 antibody to the carboxyl terminus of rhodopsin does not interfere with the Gt binding site. As shown in Fig. 2, the ability of R* in Nanodiscs to activate Gt is unaffected by antibody concentrations up to 1 µM, where >95% of the rhodopsin is expected to be bound to the antibody(30)
Figure 2.
Effect of 1D4-antibody on rhodopsin-catalyzed activation of Gt. Activation of Gt was monitored by following the binding of [35S]-GTPγS with time in the presence (diamonds) or absence (circles) of 1 µM 1D4-antibody using a filter-binding assay as described in the Experimental Procedures. Each reaction contained 5 nM of the rhodopsin triple mutant in Nanodiscs reconstituted with 11-cis-retinal, 1 µM Gt, and 3 µM GTP. Solid symbols, reaction in dark; open symbols, reaction after to exposure to light (300 W tungsten bulb for 30 sec at t = 6.5 min). Residual 1D4-peptide was present at a concentration 20-fold lower than that of the antibody combining sites.
Purification and Characterization of a R*/Gt Complex in Nanodiscs
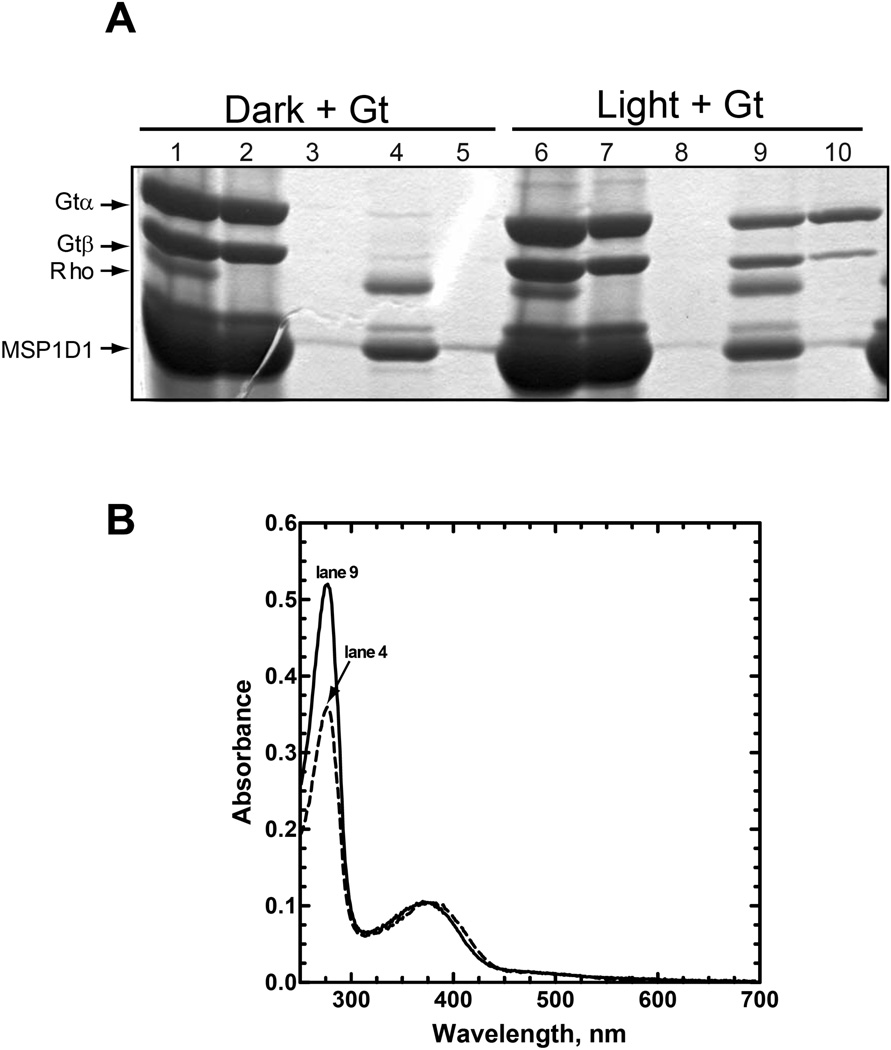
Initial attempts at preparation of the R*/Gt complex in Nanodiscs were performed using mutant rhodopsin that had been reconstituted with 11-cis-retinal in the dark. Following detergent removal, the mixture of empty and rhodopsin-containing Nanodiscs were combined with a two-fold molar excess (relative to rhodopsin) of Gt in Buffer B, incubated on ice for 15 minutes under dim-red light, and then illuminated with a 300w tungsten lamp for 2 minutes. Samples were then incubated on ice for an additional 15 min and applied to a 1D4-Sepharose immunoaffinity column for purification (Fig. 3A, lane 6). The nonbinding fraction (Fig. 3A, lane 7) typically contained visibly less Gt than was applied to the column, as well as small amounts of rhodopsin. The column was then washed sequentially with Buffer B, Buffer B containing empty Nanodiscs (50 µM) to remove nonspecifically bound Gt, and then Buffer B again (Fig. 3, lane 8) before elution of the complex by incubation with the 1D4-peptide (Fig. 3A, lane 9). Gt can be eluted alone from the column by incubation with GTPγS (Fig. 3A, lane 10). In this case, it is clear that some of the βγ subunit is retained on the column presumably as a result of nonspecific binding of the isoprenylated γ-subunit to the immobilized Nanodiscs (Fig. 3A, compare lane 9 with 10). Importantly, the ability to purify the complex is completely dependent on light activation of rhodopsin (Fig. 3A, compare lanes 9 & 10 with 4 & 5).
Figure 3.
Preparation and purification of an R*/Gt complex. Rhodopsin reconstituted with 11-cis-retinal in Nanodiscs was mixed with a 2-fold excess of Gt and then either incubated in the dark or illuminated for 2 min with light from a 300W tungsten bulb, as indicated in the figure. The mixture was then applied to a 1D4-Sepharose immunoaffinity column for purification as described in the Experimental Procedures. (A) Commassie Blue-stained SDS-PAGE gel containing samples from each step in the purification procedure. Lanes 1–5, sample in the dark; lanes 6–10, sample exposed to light. Lanes 1 & 6, material loaded onto the 1D4-Sepharose column; lanes 2 & 7, unbound material; lanes 3 & 8, last wash; lanes 4 & 9, 1D4-eluate; and lanes 5 & 10, GTPγS eluate. Only the α- and β-subunits of Gt are shown; in this and subsequent figures, the γ-subunit is not shown because it runs off the end of the gel. Molecular weights are as follows (Da): Gtα, 39,966; Gtβ, 37,377; Rho (N2C,E113Q,D282C mutant), 38,984; and MSP1D1, 24,662. The band migrating just above MSP1D1 on the gel is an impurity from the MSP1D1 preparation and has not been identified. As is noted in the text, the lower yield of the Gtβ-subunit from the GTPγS-elution, here (lane 10) and in Fig. 7, presumably reflects nonspecific binding of the more hydrophobic, isoprenylated γ-subunit to immobilized Nanodiscs on the column. (B) UV-visible absorption spectra for the purified samples from lanes 4 and 9 from Panel A, as is indicated in the figure.
The UV-visible absorption spectrum of the purified, light-activated R*/Gt complex is characterized by an A280 nm/A380 nm ratio of 5 (Fig. 3B, lane 9 spectrum). The increased absorbance at 280 nm, compared to the dark sample from Fig. 3A, lane 4 (Fig. 3B, lane 4 spectrum; A280 nm/A380 nm ratio of 3.4) is attributed to the presence of Gt in the light-activated sample. An A280 nm/A380 nm ratio of 5 is consistent with a rhodopsin/Gt molar ratio of 1/1 (based on absorption coefficients determined for Gtempty, MSP1D1, and rhodopsin; see Experimental Procedures).
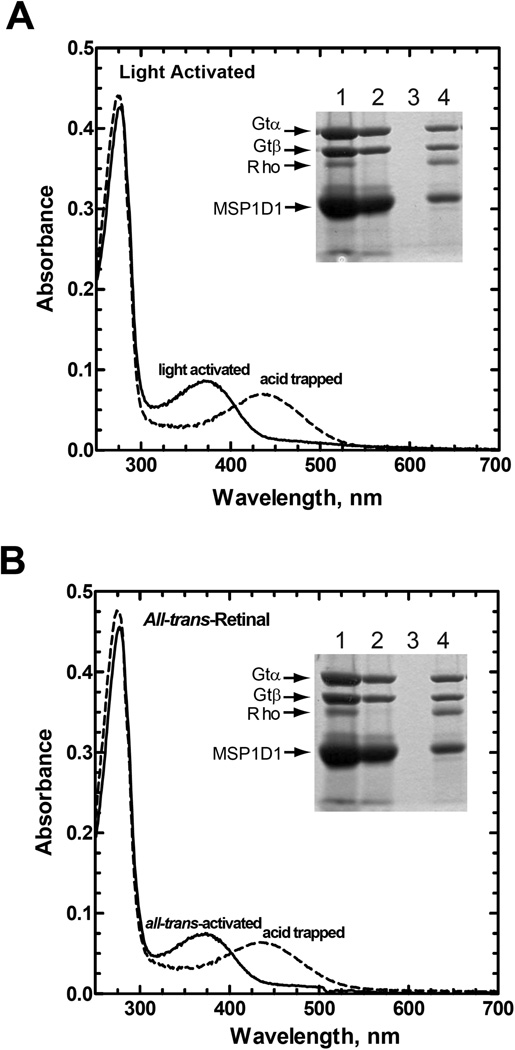
The R*/Gt complex could also be prepared from Nanodiscs containing rhodopsin that had been reconstituted and activated with all-trans-retinal (Fig. 4). The subunit composition and spectral properties of the all-trans-activated complex were very similar to those of the light-activated complex (Fig. 4, compare Panel A with B) including the ability to acid trap a protonated form of the retinylidene chromophore (Fig. 4, acid trapped spectra) demonstrating that the retinal is covalently bound in both complexes.
Figure 4.
Formation of a R*/Gt complex following activation of rhodopsin with all-trans-retinal. The figure shows absorption spectra for the purified complex before and after acidification with 50 mM sodium phosphate buffer, pH 3.5, containing 0.5% (w/v) SDS. Spectra have been corrected for dilution. Inset, SDS-PAGE analysis of the steps used for preparation and purification of the R*/Gt complexes in Nanodiscs (procedures as in Figure 3 and Experimental Procedures). Lane 1, material before loading onto the 1D4-Sepharose column; lane 2, unbound material; lane 3, last wash; and lane 4, 1D4-peptide eluate. Protein bands in the gel were visualized with Coomassie Blue. Only the α- and β-subunits of Gt are shown. A, formation of the R*/Gt complex with light-activated rhodopsin (rhodopsin sample reconstituted with 11-cis-retinal); B, formation of the complex using rhodopsin activated with all-trans-retinal.
To eliminate concern that the 1D4-antibody used in purification, which like Gt binds to the cytoplasmic surface of rhodopsin, might influence subunit stoichiometry of the complex, an alternate purification procedure was employed using ConA-Sepharose. ConA targets carbohydrate moieties on the extracellular surface of rhodopsin, opposite the binding site for Gt. Other than the affinity support and associated eluant, all other procedures for purification of the activated complex were the same as those described above using 1D4-Sepharose. As is shown in Fig. 5, the subunit composition for the activated complex purified using ConA is very similar to that purified on the 1D4-column (Fig. 4B inset). Thus, the use of the 1D4-antibody does not appear to significantly influence subunit stoichiometry in the activated complex.
Figure 5.
Purification of the R*/Gt complex using ConA-Sepharose. Rhodopsin in Nanodiscs was reconstituted with all-trans-retinal in the presence of excess Gt as described in Experimental Procedures and then purified by chromatography on ConA-Sepharose. Steps in the purification procedure were analysed on Coomassie Blue-stained SDS-PAGE gels. Lane 1, material applied to the ConA-Sepharose column; lane 2, unbound material; lane 3, last wash; and lane 4, methyl α-D-mannopyranoside eluate. Only the α- and β-subunits of Gt are shown on the gel.
Nucleotide-Binding State of Gt in the Complex
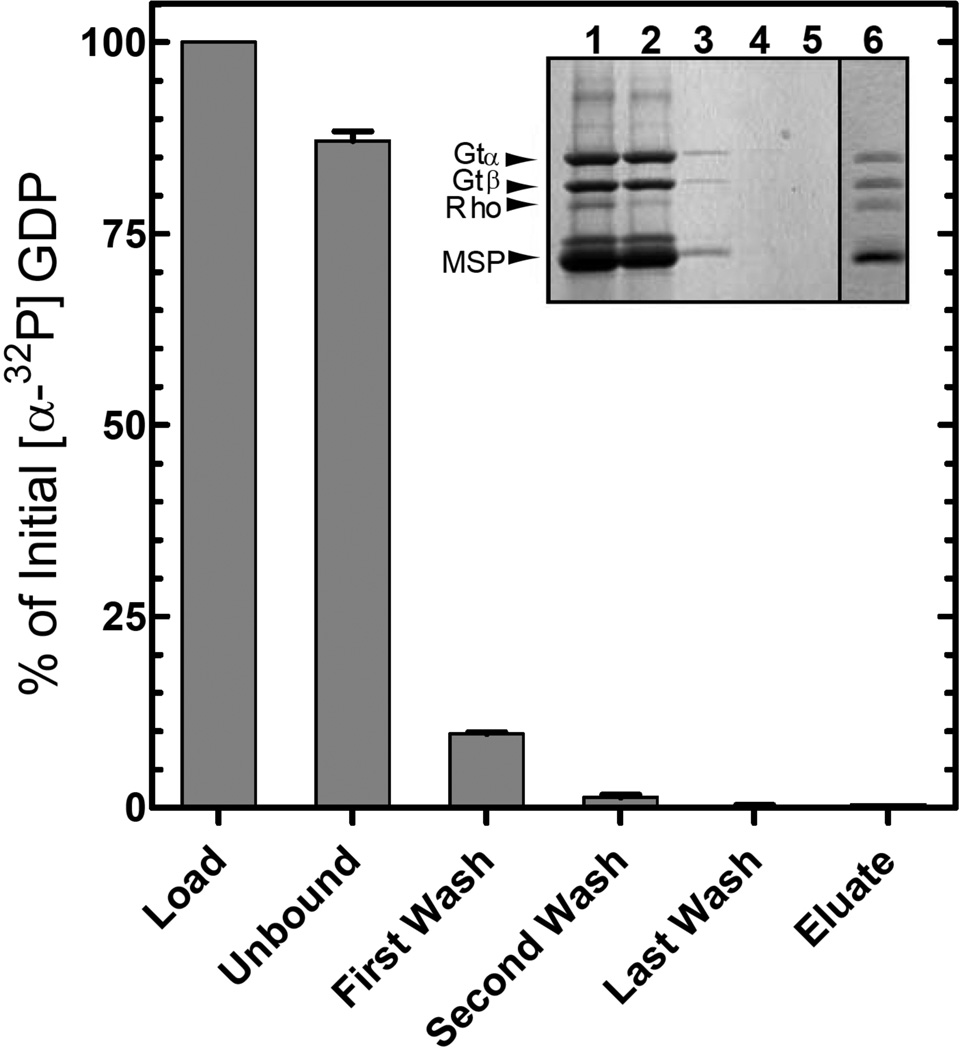
It is well established that rhodopsin catalyzes release of GDP from Gtα upon formation of the R*/Gt complex. To determine if nucleotide was released in the Nanodisc preparations, Gt loaded with [α-32P]-GDP of known specific activity in the nucleotide-binding pocket was used to form the activated complex. As is shown in Fig. 6, most of the GDP (87 ± 1.2 %, n = 3) was found in the unbound fraction from the 1D4-Sepharose column, 9.7 ± 0.3 % (n = 3) in the first wash, and none in the eluate, which contained the expected content of Gt in the complex (Fig. 6 inset, lane 6). Therefore, the nucleotide-binding pocket of Gt in the activated complex is empty.
Figure 6.
Nucleotide release during formation of the R*/Gt complex. The activated complex was purified on 1D4-Sepharose as described in Figure 3B and Experimental Procedures using a Gt sample containing [α-32P]-GDP in the nucleotide-binding pocket. Radioactivity in the fractions was monitored by liquid scintillation counting. Values are expressed as a percent of the total [α-32P]-GDP in the loaded sample. Error bars represent standard deviation with n = 2. Inset, SDS-PAGE analysis of the steps in purification of the complex as visualized by Coomassie Blue staining. Lane 1, material applied to the 1D4-Sepharose column; lane 2, unbound material; lane 3, first wash; lane 4, second wash; lane 5, last wash; and lane 6, 1D4-peptide eluate.
Stoichiometry of Subunits in the R*/Gt Complex
Initial experiments, described above, used the ratio of absorbance at 280 nm to that at 380 nm in the UV-visible absorption spectrum of the purified preparations to determine subunit composition from the known absorption coefficients for rhodopsin, transducin and MSP1D1. The amount of Gtα in the complex was also determined by simply diluting a sample with known amount of rhodopsin (from the ε380) into a reaction mixture containing 40 µM [35S]-GTPγS of known specific activity and quantifying the amount of bound GTPγS by separation of protein onto nitrocellulose filters and scintillation counting. The Gt/rhodopsin molar ratio determined by this method was 1.01 ± 0.06 (SEM, n = 3), in good agreement with the spectrophotometric determination described above.
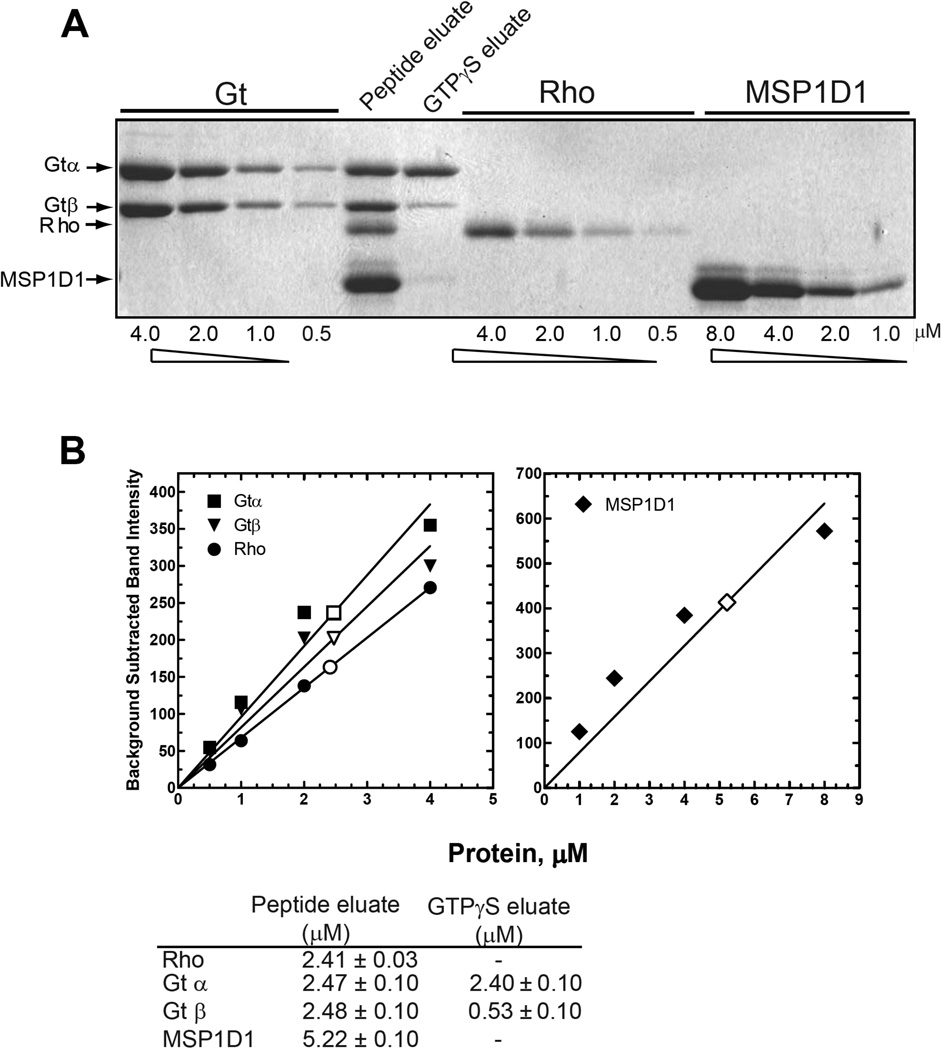
A final determination of subunit stoichiometry in the purified complex was determined by densitometry of band intensities following separation by SDS-PAGE (Fig. 7). Gels containing samples of purified complex from the 1D4-support and known standards for Gt, rhodopsin, and MSP1D1 were scanned, and bands in the digitized images were integrated. As shown in Fig. 7, the data are in support of a 1/1/2 complex of Gt, R*, and MSP1D1. Analysis of the GTPγS eluate confirmed a 78% loss of Gβ as compared to Gα of the same sample (see above).
Figure 7.
Densitometric determination of subunit stoichiometry in the R*/Gt complex. (A) SDS-PAGE analysis of the R*/Gt complex. Known amounts of Gt, the N2C,E113Q,D282C mutant (Rho), and MSP1D1 were loaded onto the gel to generate standard curves for determination of the amount of each subunit contained in the 1D4-peptide and GTPγS eluates. B, Quantification of subunits in the complex as determined by densitometry. Individual bands from the SDS-gel in A were quantified as described in Experimental Procedures using ImageJ software, and standard curves derived from the known amounts of the α- and β-subunits of Gt, rhodopsin, and MSP1D1 were generated (closed symbols) and used to determine concentrations of the subunits in the 1D4-peptide (open symbols) and GTPγS eluates (not shown), as presented in the table at the bottom of the panel. Errors were determined from least-squares fit of data to the standard curves.
DISCUSSION
While the biochemical literature on the interaction of rhodopsin and Gt is extensive and dates back to the early 1980s(4, 31–33), there remains some controversy about the stoichiometry of the activated complex of these two proteins. Rhodopsin is known to form higher-order aggregates in rod outer segment disc membranes(34), and there are reports suggesting that the functional unit is comprised of two rhodopsin molecules to one Gt(6, 7). On the other hand, the fact that rod photoreceptor cells can respond to a single photon(35) suggests that one molecule of rhodopsin is sufficient to activate Gt. In addition, there appears to be general agreement in the field that a single molecule of rhodopsin is sufficient to activate Gt in in vitro assays(8, 10, 11, 15).
We previously reported the purification of a R*/Gt complex in detergent solution using either ConA- or 1D4-Sepharose as the supporting affinity matrix(9). The complex, which was formed from the rhodopsin N2C,E113Q,D282C triple mutant and native Gt, contained a covalently bound all-trans-retinal chromophore in rhodopsin and an empty nucleotide-binding pocket in Gt. It was active, dissociated upon addition of guanine nucleotide, and was characterized by a 1/1 molar ratio of rhodopsin to Gt. A similar complex, with 1/1 molar ratio of subunits, was also purified and characterized using native rhodopsin(9). The 1/1 stoichiometry agreed well with previous studies from Ernst et al.(8) using size-exclusion chromatography to prepare the complex from native rhodopsin and Gt. Importantly, however, none of these studies unequivocally identified the number of protein subunits in the activated complex, as a molar ratio of 1/1 cannot be distinguished from 2/2, 3/3, and higher order aggregates. For this reason, we used Nanodiscs to isolate single molecules of rhodopsin. We previously reported the preparation of a R*/Gt complex in Nanodiscs using native rhodopsin but did not rigorously determine subunit stoichiometry in that study(11).
The R*/Gt complex reported here using Nanodiscs had properties that were very similar to those reported for the complex in detergent solution(9). The complex contains a covalently bound all-trans-retinal chromophore in rhodopsin and an empty nucleotide-binding pocket in Gt. It is active, dissociates upon addition of guanine nucleotide, and, most importantly, is characterized by a 1/1 molar ratio of rhodopsin to Gt. In this case, however, we may conclude that the complex is composed of 1 molecule of rhodopsin and 1 molecule of Gt because the Nanodiscs were prepared under conditions in which each contained only a single molecule of rhodopsin. This result agrees well with the subunit composition determined by x-ray crystallography for an activated complex of the β2-adrenergic receptor and Gs determined by Kobilka and coworkers; the β2-adrenergic receptor/Gs complex is composed of a single molecule of receptor bound to a single molecule of the G protein(36). It also agrees well with several studies showing that a single molecule of rhodopsin is sufficient for phosphorylation by rhodopsin kinase(12, 14) and that rhodopsin binds arrestin in a complex composed of a single molecule of rhodopsin and a single molecule of arrestin(12–14).
A comment on the use of a rhodopsin mutant in these studies is warranted. The N2C,E113Q,D282C triple mutant combines the properties of two previously described rhodopsin mutants: a double mutant, N2C,D282C, that contains an engineered disulfide bond between two introduced Cys residues(17), and a single mutant, E113Q, that neutralizes the Schiff base counterion(37, 38). The engineered disulfide confers enhance thermal stability to the protein but is otherwise without effect on activity(17). A crystal structure of this mutant in the dark state is identical to those of the native protein with the exception of the missing carbohydrate chain normally found attached to Asn 2 and the presence a disulfide bond between the two introduced Cys residues at positions 2 and 282(39). The E113Q mutation leads to a large change in the spectral properties of the dark state protein, shifting the absorption maximum from 500 nm for the retinylidene chromophore in the native protein to 380 nm in the mutant(37, 38) and leads to constitutive activation of the apoprotein opsin(16). Importantly for the current study, the mutation also prolongs the lifetime of the active metarhodopsin II state(37). The phenotypes of both mutations, N2C,D282C and E113Q, combine in the triple mutant(9), but otherwise the protein displays wild-type properties. For example, a crystal structure of the N2C,E113Q,D282C mutant in the active state superimposes with that of the WT protein(40), as does also another mutant that combines a constitutively active mutation (M257Y) with the engineered disulfide bond between residues 2 and 282(41). We previously used the N2C,E113Q,D282C triple mutant to determine the stoichiometry of the Rho*/Gt complex in detergent solution to be 1/1(9). In that study, we also showed that, under identical conditions, native rhodopsin from bovine retina and the N2C,D282C double mutant formed similar complexes of 1/1 stoichiometry with Gt. Thus, in conclusion, it is unlikely that use of the N2C,E113Q,D282C triple mutant affects stoichiometry of the activated rhodopsin/Gt complex. As a final note, we had previously prepared an activated complex of native rhodopsin and Gt in Nanodiscs containing only a single rhodopsin(11). While we did not rigorously determine stoichiometry of the complex, Coomassie-staining of bands from SDS-PAGE gels in that study(11) is consistent with the patterns reported here for the 1/1 complex with the mutant, bolstering the conclusion that use of the triple mutant does not affect the results.
While the impetus for initiating these studies was to determine subunit stoichiometry for the activated complex, our current focus is on adapting the Nanodisc system for use with total internal reflection fluorescence microscopy to follow the interaction of R* and Gt at the single molecule level. Nanodiscs would appear to be an ideal system for such studies.
ACKNOWLEDGMENTS
We gratefully acknowledge the participation of Dr. Tim Bayburt, supported in part by NIH grant GM075365, in early experiments. We thank Matt Ranaghan and Darren Mushrush for comments on the manuscript.
Funding
This work was supported by NIH grants EY007965 (D.D.O.), EY18542 (A.M.D.), and GM33775 (S.G.S.)
Abbreviations
- GDP
guanosine 5′-diphosphate
- GTP
guanosine 5′-triphosphate
- GTPγS
guanosine 5’-(γ-thio)triphosphate
- ROS
rod outer segment
- R*
activated rhodopsin
- Gt
transducin
- Gtα
α subunit of transducin
- Gtβ
β subunit of transducin
- GtGDP
transducin containing GDP
- Gtempty
transducin with no nucleotide bound
- CHAPS
3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate
- ConA-Sepharose
concanavalin A-Sepharose 4B
- DDM
dodecyl β-D-maltoside
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- HEK
human embryonic kidney
- MWCO
molecular weight cut-off
- POPC
1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine
- ROS
rod outer segments
- SDS
sodium dodecyl sulfate
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 2.Smith SO. Structure and activation of the visual pigment rhodopsin. Annu Rev Biophys. 2010;39:309–328. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- 3.Palczewski K. Chemistry and biology of vision. J Biol Chem. 2012;287:1612–1619. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emeis D, Kuhn H, Reichert J, Hofmann KP. Complex formation between metarhodopsin II and GTP-binding protein in bovine photoreceptor membranes leads to a shift of the photoproduct equilibrium. FEBS Lett. 1982;143:29–34. doi: 10.1016/0014-5793(82)80266-4. [DOI] [PubMed] [Google Scholar]
- 5.Jastrzebska B, Goc A, Golczak M, Palczewski K. Phospholipids are needed for the proper formation, stability, and function of the photoactivated rhodopsintransducin complex. Biochemistry. 2009;48:5159–5170. doi: 10.1021/bi900284x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. Faseb J. 2013;27:1572–1584. doi: 10.1096/fj.12-225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jastrzebska B, Ringler P, Palczewski K, Engel A. The rhodopsintransducin complex houses two distinct rhodopsin molecules. J Struct Biol. 2013;182:164–172. doi: 10.1016/j.jsb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci U S A. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie G, D'Antona AM, Edwards PC, Fransen M, Standfuss J, Schertler GF, Oprian DD. Preparation of an activated rhodopsin/transducin complex using a constitutively active mutant of rhodopsin. Biochemistry. 2011;50:10399–10407. doi: 10.1021/bi201126r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee S, Huber T, Sakmar TP. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 11.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 12.Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, Huang CC, Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukamoto H, Sinha A, DeWitt M, Farrens DL. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vishnivetskiy SA, Ostermaier MK, Singhal A, Panneels V, Homan KT, Glukhova A, Sligar SG, Tesmer JJ, Schertler GF, Standfuss J, Gurevich VV. Constitutively active rhodopsin mutants causing night blindness are effectively phosphorylated by GRKs but differ in arrestin-1 binding. Cell Signal. 2013;25:2155–2162. doi: 10.1016/j.cellsig.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whorton MR, Jastrzebska B, Park PS, Fotiadis D, Engel A, Palczewski K, Sunahara RK. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson PR, Cohen GB, Zhukovsky EA, Oprian DD. Constitutively active mutants of rhodopsin. Neuron. 1992;9:719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- 17.Xie G, Gross AK, Oprian DD. An opsin mutant with increased thermal stability. Biochemistry. 2003;42:1995–2001. doi: 10.1021/bi020611z. [DOI] [PubMed] [Google Scholar]
- 18.Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 19.MacKenzie D, Arendt A, Hargrave P, McDowell JH, Molday RS. Localization of binding sites for carboxyl terminal specific anti-rhodopsin monoclonal antibodies using synthetic peptides. Biochemistry. 1984;23:6544–6549. doi: 10.1021/bi00321a041. [DOI] [PubMed] [Google Scholar]
- 20.Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Expression of a Synthetic Bovine Rhodopsin Gene in Monkey Kidney Cells. PNAS. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 22.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible Nacetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wessling-Resnick M, Johnson GL. Allosteric behavior in transducin activation mediated by rhodopsin. Initial rate analysis of guanine nucleotide exchange. J Biol Chem. 1987;262:3697–3705. [PubMed] [Google Scholar]
- 24.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Fasick JI, Lee N, Oprian DD. Spectral tuning in the human blue cone pigment. Biochemistry. 1999;38:11593–11596. doi: 10.1021/bi991600h. [DOI] [PubMed] [Google Scholar]
- 26.Gasteiger E, H C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook, Humana Press. 1 ed. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 27.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 28.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhukovsky EA, Robinson PR, Oprian DD. Transducin activation by rhodopsin without a covalent bond to the 11-cis-retinal chromophore. Science. 1991;251:558–560. doi: 10.1126/science.1990431. [DOI] [PubMed] [Google Scholar]
- 30.Molday RS, MacKenzie D. Inhibition of monoclonal antibody binding and proteolysis by light-induced phosphorylation of rhodopsin. Biochemistry. 1985;24:776–781. doi: 10.1021/bi00324a036. [DOI] [PubMed] [Google Scholar]
- 31.Bornancin F, Pfister C, Chabre M. The transitory complex between photoexcited rhodopsin and transducin. Reciprocal interaction between the retinal site in rhodopsin and the nucleotide site in transducin. Eur J Biochem. 1989;184:687–698. doi: 10.1111/j.1432-1033.1989.tb15068.x. [DOI] [PubMed] [Google Scholar]
- 32.Kühn H. Interactions Between Photoexcited Rhodopsin and Light-Activated Enzymes in Rods. In: Chader J, editor. Progress in Retinal Research. Oxford, UK: Pergamon Press; 1984. pp. 123–156. [Google Scholar]
- 33.Kuhn H, Bennett N, Michel-Villaz M, Chabre M. Interactions between photoexcited rhodopsin and GTP-binding protein: kinetic and stoichiometric analyses from light-scattering changes. Proc Natl Acad Sci U S A. 1981;78:6873–6877. doi: 10.1073/pnas.78.11.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 35.Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. The Journal of physiology. 1979;288:613–634. [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhukovsky EA, Oprian DD. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 39.Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GF. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Standfuss J, Edwards PC, D'Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, Schertler G, Standfuss J. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci U S A. 2012;109:119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]