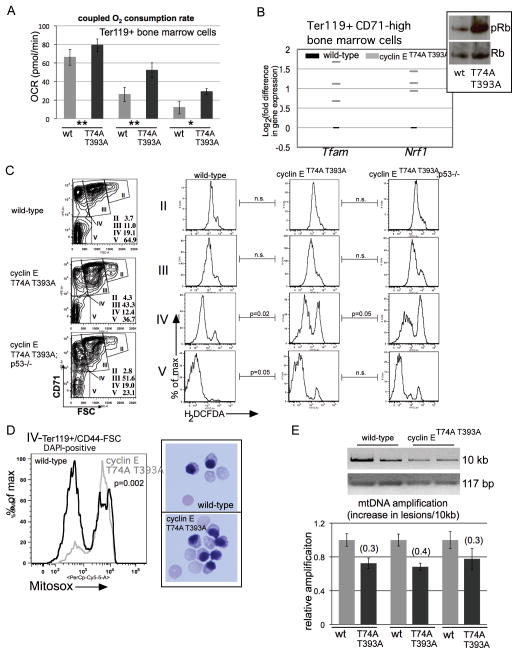
Figure 4. Oxidative metabolism and mitochondrial ROS are increased in cyclin ET74A T393A erythroid cells.
(A) Oxidative consumption rates coupled to mitochondrial ATP production were assayed in primary Ter119+ cells from three sets of mice of the indicated genotypes using a Seahorse Extracellular Flux Analyzer. (wt= wild-type, T74A T393A= cyclin ET74A T393A cells; ** - 4×105, * - 3×105 cells analyzed in independent assays). Error bars indicate standard error with four replicates per sample. (B) Expression of the indicated genes in purified Ter119+ CD71-high erythroid cells was measured by RT-PCR. Each grey bar represents averaged value of triplicate assays from a single knock-in mouse, expressed relative to expression in cells from age- and sex-matched wild-type controls. Inset shows total and phospho-Rb expression from similarly isolated cells primary erythroid cells. (C) Left column - bone marrow Ter119+ cells were subdivided into distinct morphologic subpopulations (II–V) based on CD71 expression vs. forward scatter (FSC)14, with relative abundances indicated. Right columns - intracellular reactive oxygen species were measured in the indicated erythroid cell subsets using cell-permeable 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), enabling detection by flow cytometry. Comparisons between mean fluorescence signals for bone marrow erythroid cells of four mice from each of the indicated genotype groups were made and p-values calculated using paired t-tests. (D) Left – Representative comparison is shown of mitochondrial superoxide levels measured in bone marrow erythroid cells from three pairs of mice at the indicated stage of maturation based on CD44/FSC, using Mitosox Red superoxide indicator. Right –100x micrographs of May-Grunwald/Giemsa-stained orthochromatic erythroblasts isolated using the gating employed for the Mitosox assays. (E) Top – ethidium bromide staining of the indicated mitochondrial DNA (mtDNA) amplification products, resolved on agarose gels, is shown. The mtDNA templates were obtained from Ter119+ bone marrow cells isolated from two mice of each of the indicated genotypes. Bottom – quantification of Ter119+ cell mtDNA amplification for three pairs of mice, performed using PicoGreen reagent, expressed relative to wild-type mtDNA amplification. Derivation of the calculation of for the increase in mtDNA lesions per 10kb is previously described,26 using normalization to 117 base-pair (bp) mtDNA amplification product for mitochondrial copy number.