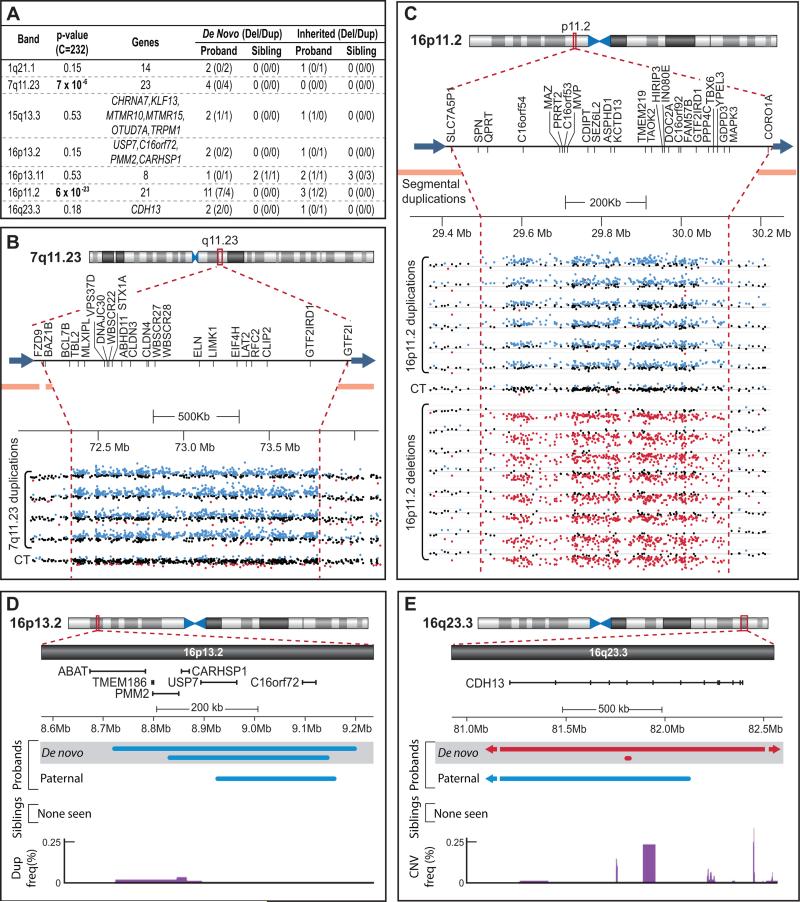
Figure 4. Confirmed recurrent rare de novo CNVs.
A) All recurrent de novo CNVs identified in 1,124 probands and 872 siblings. The gene count is given when >6 RefSeq genes map to an interval; a complete listing of genes is presented in Table S4. The total number of de novo and matching inherited CNVs in probands and siblings are shown for deletions (Del) and duplications (Dup) in parentheses. B) LogR data for 4 de novo duplications and 1 control with no CNV (CT) in the 7q11.23 interval. RefSeq genes within this region are noted below the ideogram; the orange bars represent flanking segmental duplications. NCBI 36 (hg18) genomic coordinates are shown with the scale indicated. The LogR for all probes within the region is shown; LogR values greater than 0.15 are in blue (suggesting a duplication) while LogR values less than −0.15 are in red (suggesting a deletion). B allele frequency data is not shown but supports the presence of a corresponding CNV. The approximate boundaries of the CNVs are shown by the vertical dashed red line and blue arrows. C) LogR data for 6 duplications (4 de novo), 8 deletions (7 de novo), and 1 control with no CNV (CT) in the 16p11.2 interval. The ideogram and intensity plots are as in B. D) Overlapping rare de novo and rare inherited CNVs identified in the 16p13.2 interval. The brackets show the boundaries of RefSeq genes; 2 genes are in common between all 3 duplications: USP7 and C16orf72. The frequency of duplications in the DGV is shown in purple: the majority of the recurrent de novo region is not present in the DGV. E) Overlapping rare de novo and rare inherited CNVs identified in the 16q23.3 interval. A 34kb deletion overlaps a 5Mb deletion over a CDH13 exon (represented by ticks on the gene). The frequency of CNVs observed in the DGV is shown at the bottom in purple.