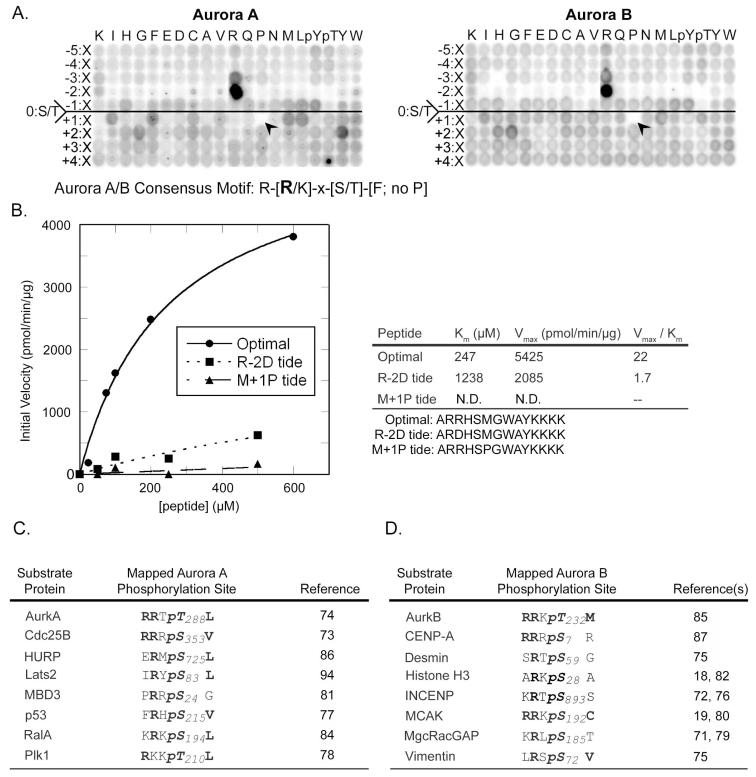
Fig. 4. Phosphorylation motif selectivity of Aurora A and B.
(A) PS-OPLS blots of Aurora A (left) and Aurora B:INCENP complex (right) reveal positively and negatively selected residues for motifs recognized by these kinases. The arrowheads identify the complete selectivity against Pro at position Ser/Thr+1. (B) Peptide phosphorylation and determination of kinetic parameters (Km, Vmax, and Vmax/Km ratio) for reactions of Aurora B:INCENP with the optimal substrate peptide and peptides with single amino acid substitutions. A graph of reaction data fitted to the Michaelis-Menten equation for peptides is indicated on the left, and a table of kinetic parameters is shown on the right. R-2D tide and M+1P tide sequences are indicated below the table with the substituted positions shown in bold. (C) Previously published phosphorylation sites on mapped substrates of Aurora A (left) and Aurora B (right) conform to the optimal phosphorylation motif determined by PS-OPLS. Bold represents motif selected residues.