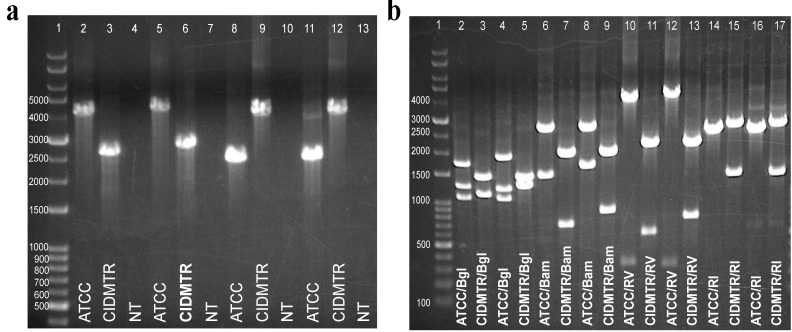
Figure 6.
Strain-specific PCR assays differentiate ATCC 22122 strain of GPCMV from CIDMTR strain. (a) Primers were designed to amplify across discontinuous/unique regions as demonstrated in Figure 3b. Lane 1, kb ladder; lane 2, mismatch region 1 F1/R1, ATCC; lane 3, mismatch region 1, F1/R1, CIDMTR; lane 4, mismatch region 1, no template control; lane 5, mismatch region 1 F2/R2, ATCC; lane 6, mismatch region 1 F2/R2, CIDMTR; lane 7, mismatch region 1 F2/R2, no template control; lane 8, mismatch region 2 F1/R1, ATCC; lane 9, mismatch region 2 F1/R2, CIDMTR; lane 10, mismatch region 2 F1/R1, no template control; lane 11, mismatch region 2 F2/R2, ATCC; lane 12, mismatch region 2 F2/R2, CIDMTR; lane 13, mismatch region 2 F2/R2, no template control. (b) Restriction polymorphisms were as predicted by DNA sequence. PCR amplification products were digested with enzymes as indicated. Lane 1, kb ladder; lane 2, mismatch region 1 F1/R1 ATCC, Bgl 2; lane 3, mismatch region 1 F1/R1 CIDMTR, Bgl 2; lane 4, mismatch region 1 F2/R2 ATCC, Bgl 2; lane 5, mismatch region 1 F2/R2 CIDMTR, Bgl 2; lane 6, mismatch region 1 F1/R1 ATCC, BamH I; lane 7, mismatch region 1 F1/R1 CIDMTR, BamH I; lane 8, mismatch region 1 F2/R2 ATCC, BamH I; lane 9, mismatch region 1 F2/R2, BamH I; lane 10, mismatch region 1 F1R1 ATCC, EcoR V; lane 11, mismatch region 1 F1/R1 CIDMTR, EcoR V; lane 12, mismatch region 1 F2/R2 ATCC, EcoR V; lane 13, mismatch region 1 F2/R2, EcoR V; lane 14, mismatch region 2 F1R1 ATCC, EcoR I; lane 15, mismatch region 2 F1R1 CIDMTR, EcoR I; lane 16, mismatch region 2 F2/R2 ATCC, EcoR I; lane 17, mismatch region 2 F2/R2, EcoR I.