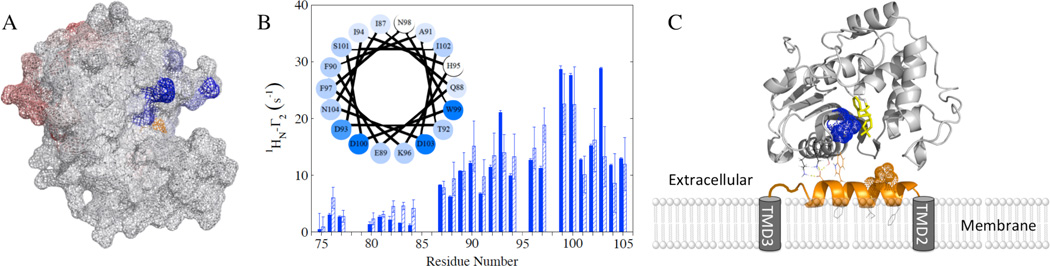
Figure 2.
(A) BlaRS PRE Γ2 rates caused by spin-labeled L2short T92C mapped onto the BlaRS crystal structure (PDB 3Q7V, chain B). Blue (β5–β6 turn) and red (β6–β7 turn) indicate the two interaction regions. Darker shading indicates larger 1HN-Γ2. The antibiotic-binding-site residue S389 is in orange. (B) L2short PRE 1HN-Γ2 rates caused by IAP-labeled BlaRS I531C without (dark blue) and with (dashed) penicillin G. The helical wheel includes I87-N104 of L2short, with darker shading for larger 1HN Γ2 rates. (C) Model for L2 interaction with BlaRS, based on PREs from I531C (BlaRS) and T92C (L2short), and CSPs from the β5–β6 turn. Coloring: BlaRS (gray), L2short helix (orange helix), I531 (blue spheres), acylation site (penicillin G in yellow sticks).