Abstract
Background
The human fetus is able to mount a systemic inflammatory response when exposed to microorganisms. This stereotypic response has been termed the “fetal inflammatory response syndrome” (FIRS), defined as an elevation of fetal plasma interleukin-6 (IL-6). FIRS is frequently observed in patients who delivered preterm associated with intra-amniotic infection (IAI), acute inflammatory lesions in the placenta, and a high rate of neonatal morbidity. Recently, a novel form of fetal systemic inflammation, characterized by an elevation of fetal plasma CXCL10, has been identified in patients with placental lesions suggestive of “maternal anti-fetal rejection”. These lesions include chronic chorioamnionitis, plasma cell deciduitis and villitis of unknown etiology (VUE). In addition, a seropositivity for HLA panel-reactive antibodies (PRA) in maternal sera can also be used as an index of suspicious for “maternal anti-fetal rejection”. The purpose of this study was to determine: 1) the frequency of pathologic evidence of “maternal anti-fetal rejection” in term and spontaneous preterm births; 2) the fetal serum concentration of CXCL10 in patients with and without evidence of maternal anti-fetal rejection; and 3) the fetal blood transcriptome and proteome in pregnancy with evidence of fetal inflammatory response associated with maternal anti-fetal rejection.
Methods
Maternal and fetal sera were obtained from normal term birth (N=150) and spontaneous preterm births (N=150). Fetal inflammatory response associated with maternal anti-fetal rejection was diagnosed when the patients met two or more of the following criteria: 1) presence of chronic placental inflammation; 2) ≥80% of maternal HLA class I panel-reactive antibody (PRA) seropositivity; and 3) fetal serum CXCL10 concentration > 75th percentile of normal. Maternal HLA PRA was analyzed by flow cytometry. The concentration of fetal CXCL10 and IL-6 were determined by ELISA. Transcriptome analysis was undertaken after extraction of total RNA from white blood cells with a whole-genome DASL assay. Proteomic analysis of fetal serum was conducted by two-dimensional difference gel electrophoresis. Differential gene expression was considered significant when there was a p<0.01 and a fold-change >1.5.
Results
1) The frequency of placental lesions consistent with maternal anti-fetal rejection was higher in patients with preterm delivery than in those with term delivery (56% vs. 32%; P<0.001); 2) patients with spontaneous preterm births had a higher rate of maternal HLA PRA class I positivity than those who delivered at term (50% vs. 32%; P=0.002); 3) fetuses who were born to mothers with positive maternal HLA PRA results had a higher median serum CXCL10 concentration than in those with negative HLA PRA results (P<0.001); 4) the median serum CXCL10 concentration (but not IL-6) was higher in fetuses with placental lesions associated with maternal anti-fetal rejection than in those without such lesions (P<0.001); 5) a whole-genome DASL assay of fetal blood RNA demonstrated differential expression of 128 genes between fetuses with and without fetal inflammatory response associated with maternal anti-fetal rejection; and 6) comparison of the fetal serum proteome demonstrated 20 proteins whose abundance differed between fetuses with and without fetal inflammatory response associated with maternal anti-fetal rejection.
Conclusions
We describe systemic inflammatory response in the fetus born to mothers with evidence of maternal anti-fetal rejection. Using high-dimensional biology techniques, the transcriptome and proteome of this novel type of fetal inflammatory response demonstrated the distinct profile from FIRS type I (which is associated with acute infection). This information is crucial to gain a mechanistic understanding of the syndrome as well as to identify biomarkers for this condition.
Keywords: anti-HLA panel-reactive antibody, apolipoprotein C-III, CD34, CXCL10, chronic placental inflammation, pregnancy, proteome, transcriptome
Introduction
Pregnancy is a unique immunologic state in which the maternal adaptive and innate components of the immune system support the establishment and maintenance of pregnancy, and provide defense mechanisms against microbial pathogens.1,2 The fetus is a semi-allograft, and active maternal immune tolerance mechanisms are fundamental for a tolerogenic state of paternal antigens and the prevention of the rejection of the fetus. 1,3–25
The diagnosis of maternal anti-fetal rejection has been a challenge to clinical obstetrics and surgical pathology. We have recently reported a series of studies demonstrating that maternal anti-fetal rejection can be a mechanism of disease associated with spontaneous preterm birth and can be diagnosed by the identification of chronic chorioamnionitis, a lesion characterized by maternal T-cell infiltration of the chorioamniotic membranes.26–29 Other pathologic lesions reflecting maternal anti-fetal rejection included chronic deciduitis with plasma cells and villitis of unknown etiology (VUE).30
Given the unique anatomical relationship between the mother and fetus, maternal anti-fetal cellular rejection and antibody-mediated rejection can affect the fetus by mechanisms operative in graft-versus-host disease (GVHD) and alloimmune reactions.27,30 Specifically, maternal antibodies against paternal antigens can cross the placenta to activate complement and elicit a fetal inflammatory response.27
We have previously reported that the fetal plasma concentration of CXCL10 is higher for cases in which the placenta has VUE.30 Therefore, we hypothesized that maternal anti-fetal rejection is linked to a stereotypical derangement of the systemic fetal chemokine milieu, specifically CXCL10, just as intra-amniotic infection/inflammation is associated with an elevation of the fetal plasma concentration of IL-6.31–35 The latter condition observed in human fetuses of patients with preterm labor and preterm prelabor rupture of membranes (PPROM) has been termed the “fetal inflammatory response syndrome” (FIRS),31–33,36–44 and has been associated with a higher rate of adverse neonatal outcome,31–36,45–50 a short interval to delivery, and multi-systemic involvement.32,42–44,47,51–118 We have recently provided evidence that an elevation of amniotic fluid CXCL10 concentration during the mid-trimester is a risk factor for preterm delivery after 32 weeks of gestation,119 while an elevation of amniotic fluid IL-6 concentration is associated with preterm delivery before 32 weeks of gestation.119 This observation suggests that there is heterogeneity in the nature of the intra-amniotic inflammatory response during pregnancy.119 Typically, an elevation of amniotic fluid IL-6 is observed in cases of intra-amniotic infection associated with acute chorioamnionitis and funisitis 33,120–126. CXCL10 is a T-cell chemokine which is elevated in the amniotic fluid of patients with chronic chorioamnionitis.26,27,127 In this lesion, maternal T cells invade the chorioamniotic membranes,26 presumably because of the chemotactic gradient generated in the amniotic cavity by T-cell chemokines including CXCL10.26,27,127
This study was conducted to determine: 1) the frequency of pathologic evidence of cellular and humoral maternal anti-fetal rejection in term and spontaneous preterm births; 2) the fetal plasma concentration of CXCL10 in patients with and without evidence of maternal anti-fetal rejection; and 3) the fetal blood transcriptome and proteome in patients with fetal inflammatory response associated with maternal anti-fetal rejection.
Materials and Methods
Patients and definitions
The patient population comprised Hispanic women who were enrolled and delivered at the Sótero del Río Hospital, Santiago, Chile. Sera and tissue samples from the patients and their singleton neonates were retrieved from the Bank of Biological Materials of the Sótero del Río Hospital and the Perinatology Research Branch, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U. S. Department of Health and Human Services.
Patients included women who delivered (1) with a normal pregnancy outcome at term (N=150) and (2) before 37 completed weeks of gestation after preterm labor with intact membranes or PPROM (N=150). Pregnancies with a fetal congenital anomaly and small-for-gestational-age neonate were ineligible to participate. Placental tissues and fetal cord blood samples were collected at the time of delivery. We selected maternal blood samples which were collected within seven days before and after delivery to maintain a meaningful temporal relationship between placental histopathologic findings and concentrations of CXCL10 and IL-6 in maternal sera. Samples were stored at −80°C until use. All patients provided written informed consent at the Sótero del Río Hospital. The Institutional Review Boards of the participating institutions approved the collection and use of biological materials and clinical data for research purposes.
Preterm labor was defined as the presence of regular uterine contractions occurring at a frequency of at least two every 10 minutes associated with cervical dilatation, followed by delivery before 37 completed weeks of gestation. PPROM was diagnosed by sterile speculum examination when pooling of amniotic fluid in the vagina occurred or when positive nitrazine and ferning tests, conducted when necessary, were confirmed before 37 completed weeks of gestation in the absence of labor.
Placental Pathology
Placental histopathologic changes were defined according to diagnostic criteria proposed by the Perinatal Section of the Society for Pediatric Pathology and included lesions consistent with amniotic fluid infection, maternal vascular underperfusion, and fetal vascular thrombo-occlusive disease.128 The diagnosis of VUE was based on histologic criteria previously defined,30,129 and chronic chorioamnionitis was diagnosed when lymphocytic infiltration into the chorionic trophoblast layer or chorioamniotic connective tissue was present as previously described. 26–28,127,130,131 Chronic deciduitis with plasma cells was defined as the presence of lymphoplasmacytic infiltration into the decidua of the basal plate.132 Chronic placental inflammation was defined upon observation of one or more findings among chronic chorioamnionitis, VUE, and chronic deciduitis with plasma cells.
Flow Cytometry for HLA Panel-Reactive Antibodies
Flow cytometric analyses of HLA class I and class II PRA in maternal sera were conducted using the FlowPRA®-I Screening Test and the FlowPRA®-II Screening Test (One Lambda, Inc., Canoga Park, CA, USA), according to the manufacturer’s instructions. HLA class I or class II microbeads were mixed with 20 μL of serum, followed by incubation for 30 min at room temperature with gentle rotation. After the microbeads were washed 3 times with 1 mL of FlowPRA® Wash Buffer by centrifugation at 9,000xg for 2 min, they were incubated with 100 μL of FITC-conjugated F(ab)2 fragment of Fcγ fragment specific goat anti-human IgG for 30 min. Thereafter, the microbeads were washed twice with 1 mL of wash buffer, and 0.5 mL of fixing solution (PBS with 0.5% formaldehyde) was added. The FL1 fluorescence of 5,000 events was analyzed using the BD™ LSR II Flow Cytometer (BD Biosciences, San Jose, CA, USA). A sample with panel-reactivity of 10% or more was considered PRA-positive.133,134
Enzyme-Linked Immunosorbent Assays for IL-6 and CXCL10
Serum concentrations of IL-6 (Human IL-6 Quantikine® HS ELISA Kit, R&D Systems, Minneapolis, MN, USA) and of CXCL10 (Human CXCL10/IP-10 Quantikine® ELISA Kit, R&D Systems) were measured with specific immunoassays, according to the manufacturer’s instructions.
Whole-Genome DASL Assay
To characterize the fetal blood transcriptome in patients with evidence of fetal inflammatory response associated with maternal anti-fetal rejection, the Whole-Genome DASL® Assay (cDNA-mediated Annealing, Selection, Extension, and Ligation: Illumina, Inc., San Diego, CA, USA) was performed using fetal blood samples from cases with (N=9) and without (N=15) evidence of fetal inflammatory response associated with maternal anti-fetal rejection which was defined as the presence of two or more of the following criteria: 1) chronic placental inflammation (villitis of unknown etiology, chronic chorioamnionitis or chronic deciduitis with plasma cells), 2) ≥80% of maternal HLA class I PRA seropositivity; and 3) fetal serum CXCL10 concentration > 75th percentile. Group 1 comprised cases with evidence of fetal inflammatory response associated with maternal anti-fetal rejection, and nine neonates (five term and four preterm births) met the criteria. In Group 2 (cases without evidence of fetal inflammatory response associated with maternal anti-fetal rejection), 15 neonates (seven term and eight preterm births) had no chronic placental inflammation, negative maternal HLA class I PRA (<10% of panel-reactivity), and fetal CXCL10 concentration less than the 25th percentile.
Fetal cord blood samples were collected into PAXgene™ Blood RNA collection tubes (PreAnalytiX GmbH, Hombrechtikon, Switzerland). Blood tubes were kept at room temperature for 24 h and then frozen at −70°C until assay. Total blood RNA was isolated using the PAXgene™ Blood RNA Kit (Qiagen, Valencia, CA, USA) with DNase I treatment. The quantity and quality of RNA were evaluated by the Dropsense96® Microplate Spectrophotometer (Trinean, Gentbrugge, Belgium) and the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), respectively.
Gene expression of whole blood RNA was measured using the Whole-Genome DASL® Assay. One hundred nanograms of total RNA were reverse-transcribed with biotin-labeled oligo-dT and random primers. Biotinylated cDNAs were annealed to assay-specific oligonucleotides [DASL® Assay Pool (DAP) probe groups]. The mixtures were then bound on streptavidin-conjugated paramagnetic particles to select the cDNA/oligo complexes. PCR amplification was completed with fluorescently labeled primers, and the amplified PCR products were hybridized overnight onto the BeadChips (Illumina). The intensities of fluorescence were measured using the iScan™ System (Illumina).
Raw gene expression levels were normalized using the quantile normalization method.135 A linear model was used to fit gene expression levels as a function of disease status (cases with and without evidence of fetal inflammatory response associated with maternal anti-fetal rejection), gestational age at delivery (term or preterm), and gender of the fetus. Coefficients were calculated using moderated t-tests.136 Differential gene expression was considered significant based upon two criteria: a) the P value of <0.01 and b) the magnitude of change (fold-change >1.5).137 Gene Ontology analysis was conducted using an over-representation approach previously described138 and implemented in the GOstats package.139
The DASL® Assay data used in this study were submitted to the Gene Expression Omnibus (GEO). Interested readers can use the following link to access the data: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=fpwjrqimaqgeehi&acc=GSE28387.
The quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) assay was conducted to confirm DASL® Assay results for genes of interest using the Biomark™ System (Fluidigm, South San Francisco, CA, USA) with specific TaqMan® assays (Applied Biosystems®, Life Technologies Corporation, Foster City, CA, USA), according to the manufacturers’ instructions (Supplemental Table I).
Two-dimensional Difference Gel Electrophoresis (2D-DIGE)
An equal amount of fetal serum samples obtained from cases with (N=10) and without (N=10) evidence of fetal inflammatory response associated with maternal anti-fetal rejection was pooled to compare their proteome. The presence and absence of fetal inflammatory response associated with maternal anti-fetal rejection were defined by the same criteria used in the Whole-Genome DASL® assay; fetal serum CXCL10 concentration for only one case with evidence of fetal inflammatory response associated with maternal anti-fetal rejection was higher than the 50th but less than the 75th percentile. For each sample, 5 μL of lysis buffer [30 mM Tris-HCl (pH 8.8), 7 M urea, 2 M thiourea, 4% CHAPS] were added to 1 μL of serum, followed by labeling with Cy3 or Cy5. The labeling reaction was stopped by adding 1 μL of 10 mM Lysine to each pooled sample, followed by incubation on ice in the dark for an additional 15 min. Labeled samples were then mixed with 2X sample buffer [8 M urea, 4% CHAPS, 20 mg/mL dithiothreitol (DTT), 2% pharmalytes] and 100 μL of DeStreak Rehydration Solution (7 M urea, 2 M thiourea, 4% CHAPS, 20 mg/mL DTT, 1% pharmalytes; GE Healthcare Life Sciences, Piscataway, NJ, USA) for a total volume of 250 μL. The samples were mixed, spun, and then loaded into a strip holder. After isoelectric focusing (pH 3-10), IPG strips were incubated in equilibration buffer-1 (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 10 mg/mL DTT) for 15 min with gentle shaking, and rinsed in equilibration buffer-2 [50 mM Tris-HCl (pH 8.8), 6 M urea, 30% glycerol, 2% SDS, 45 mg/mL DTT] for 10 min with gentle shaking. Following electrophoresis in a 12% SDS-polyacrylamide gel at 15°C, the gel was scanned using Typhoon Trio™ (GE Healthcare Life Sciences). Scanned images were then analyzed by ImageQuant TL software version 6.0 (GE Healthcare Life Sciences), followed by differential in-gel analysis using DeCyder™ 2D Software Version 6.5 (GE Healthcare Life Sciences), to obtain the fold-changes of protein expression.
Mass Spectrometry
Twenty spots of interest were picked up by the Ettan™ Spot Picker (GE Healthcare Life Sciences) and digested in gel with modified porcine trypsin protease (Trypsin Gold; Promega, Madison, WI, USA). Digested tryptic peptides were desalted by Zip-tip C18 (Millipore Corporation, Billerica, MA, USA). Peptides were eluted from the ZipTips® with 0.5 μL of matrix solution (5 mg/mL of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 0.1% trifluoroacetic acid, and 25 mM of ammonium bicarbonate) and spotted on the MALDI plate. MALDI-TOF and TOF/TOF mass spectrometry were performed with an AB SCIEX TOF/TOF™ 5800 System (AB SCIEX, Framingham, MA, USA). MALDI-TOF mass spectra were acquired in the reflectron-positive ion mode, averaging 4000 laser shots per spectrum. TOF/TOF mass spectrometry fragmentation spectra were acquired for each sample, averaging 4000 laser shots per fragmentation spectrum on each of the 10 most abundant ions present in each sample. Both the resulting peptide mass and the associated fragmentation spectra were submitted to a GPS Explorer™ Workstation equipped with a MASCOT search engine (Matrix Science Ltd., London, UK) to search the redundant database of the National Center for Biotechnology Information (NCBI). Searches were performed without constraining protein molecular weight or isoelectric point, with variable carbamidomethylation of cysteine and oxidation of methionine residues, and with one missed cleavage also allowed in the search parameters. Candidates with either a protein score of C.I.% or Ion C.I.% >95 were considered significant.
To confirm 2D-DIGE results for proteins of interest, serum concentrations of apolipoprotein E and apolipoprotein C-III were measured with specific immunoassays (Human Apolipoprotein E ELISA Kit, Kamiya Biomedical Company, Seattle, WA, USA; AssayMax Human Apolipoprotein C-III ELISA Kit, AssayPro LLC, St. Charles, MO, USA), according to the manufacturers’ instructions.
Statistical Analysis
To obtain statistical significance for continuous variables, distributions were examined for normality using the Kolmogorov-Smirnov test. When data were far from normality, the Kruskal-Wallis one-way analysis of variance and the Mann-Whitney U tests were performed. When there was normality of continuous variables, the one-way ANOVA test and unpaired t-tests were used to compare differences. To assess the categorical variables, proportions were compared with Fisher’s exact test or the χ2 test. Medians and inter-quartile ranges were reported for continuous variables whereas frequencies and percentages were calculated for categorical variables. The Jonckheere-Terpstra test was used to compare continuous variables among multiple-ordered groups, and the linear-by-linear association analysis was used for categorical variables. Statistical analyses were performed using the SPSS Version 15.0 (SPSS, Inc., Chicago, IL, USA). All P values were two-sided, with P<0.05 considered statistically significant.
Results
Demographics of the Study Population
Table I showed the clinical characteristic and pathologic findings of the placenta and HLA PRA positivity in patients who delivered at term as well as those who had spontaneous preterm births. Histological evidence of maternal anti-fetal cellular rejection in the placenta (chronic chorioamnionitis, VUE, or chronic deciduitis with plasma cells) was more common in patients with spontaneous preterm delivery than in those who delivered at term [56% (84/150) of spontaneous preterm and 32% (48/150) of term births; P<0.001]. Maternal HLA class I PRA positivity was more common in spontaneous preterm births than in term deliveries [50% (75/150) versus 32% (48/150); P=0.002].
Table I.
Demographics and clinical characteristics of the study population
| Term delivery | Spontaneous preterm delivery | ||
|---|---|---|---|
| n=150 | n=150 | ||
| Maternal age (year)* | 27 (17–43) | 25 (15–44) | NS |
| Gestational age at delivery (weeks)* | 39.5 (37.0–41.6) | 34.8 (22.9–36.9) | <0.001 |
| Birth weight (g)* | 3440 (2650–4110) | 2460 (530–3900) | <0.001 |
| Baby gender (male, %) | 54.7 (82/150) | 66.7 (100/150) | 0.033 |
| Cesarean delivery (%) | 50.0 (75/150) | 22.0 (33/150) | <0.001 |
| Primigravida (%) | 16.0 (24/150) | 38.7 (58/150) | <0.001 |
| Nullipara (%) | 18.0 (27/150) | 42.7 (64/150) | <0.001 |
| Cellular rejection | |||
| CCA (%) | 14.0 (21/150) | 44.0 (66/150) | <0.001 |
| VUE (%) | 16.7 (25/150) | 22.0 (33/150) | NS |
| CDP (%) | 14.7 (22/150) | 26.0 (39/150) | 0.015 |
| One or more of CCA/VUE/CDP (%) | 32.0 (48/150) | 56.0 (84/150) | <0.001 |
| Severity of chronic inflammation (%) | <0.001 | ||
| None of CCA/VUE/CDP (%) | 68.0 (102/150) | 44.0 (66/150) | |
| One of CCA/VUE/CDP (%) | 22.7 (34/150) | 28.0 (42/150) | |
| Two of CCA/VUE/CDP (%) | 5.3 (8/150) | 20.0 (30/150) | |
| All of CCA/VUE/CDP (%) | 4.0 (6/150) | 8.0 (12/150) | |
| Humoral rejection | |||
| Maternal HLA class I PRA positive (%)† | 32.0 (48/150) | 50.0 (75/150) | 0.002 |
| Maternal HLA class II PRA positive (%)† | 18.0 (27/150) | 18.7 (28/150) | NS |
| Fetal HLA class I PRA positive (%)† | 18.0 (27/150) | 22.7 (34/150) | NS |
| Fetal HLA class II PRA positive (%)† | 8.7 (13/150) | 2.7 (4/150) | 0.025 |
Median (range).
Positive HLA PRA is defined as 10% or more of reactivity of HLA panel-reactive antibodies.
ACA, acute chorioamnionitis; CCA, chronic chorioamnionitis; CDP, chronic deciduitis with plasma cells; HLA, human leukocyte antigen; NS, not significant; PRA, panel-reactive antibodies; VUE, villitis of unknown etiology.
Anti-fetal Cellular Rejection and Fetal Blood CXCL10 Concentration
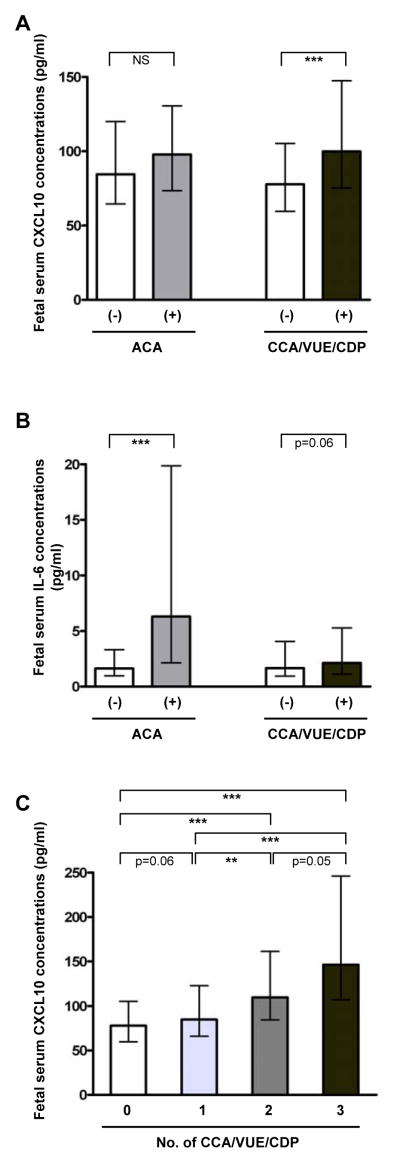
The median fetal serum CXCL10 concentration was higher in cases with anti-fetal cellular rejection than in those without cellular rejection (median 99.9 pg/mL, interquartile range [IQR] 75.2–147.5 pg/mL versus median 77.7 pg/mL, IQR 59.6–105.4 pg/mL P<0.001), while there was no difference in fetal serum CXCL10 concentration in the presence or absence of acute chorioamnionitis (Fig. 1A). Differences in fetal serum CXCL10 concentration according to the presence or absence of each type of anti-fetal cellular rejection remained significant (for chronic chorioamnionitis: median 99.0 pg/mL, IQR 75.7–147.5 pg/mL versus median 81.2 pg/mL, IQR 62.0–115.8 pg/mL; for VUE: median 128.5 pg/mL, IQR 93.9–181.2 pg/mL versus median 80.6 pg/mL, IQR 61.7–112.2 pg/mL; for chronic deciduitis with plasma cells: median 115.8 pg/mL, IQR 82.2–161.2 pg/mL versus median 81.2 pg/mL, IQR 63.3–114.3 pg/mL) (P<0.01, for each). In contrast, median fetal serum IL-6 concentrations were different between cases with and without acute chorioamnionitis (P<0.001), while there was a tendency toward higher fetal serum IL-6 concentration in those with anti-fetal cellular rejection (P=0.06) (Fig. 1B). The fetal serum concentration CXCL10 was correlated to the extent of the cellular rejection (aggregate number of pathologic lesions consistent with maternal anti-fetal rejection) (P<0.001; Fig. 1C).
Figure 1. Fetal serum CXCL10 and IL-6 concentrations according to the presence or absence of maternal anti-fetal cellular rejection.
(A) Fetal serum CXCL10 concentration was higher in cases with anti-fetal cellular rejection (chronic placental inflammation) than in those without (P<0.001), while fetal serum CXCL10 concentration was not different according to the presence or absence of acute chorioamnionitis. (B) Cases with acute chorioamnionitis had higher median fetal serum IL-6 concentration than those without (P<0.001), and fetal serum IL-6 concentration tended to be higher in cases with anti-fetal cellular rejection than in those without (P=0.06). (C) The upward trend of blood CXCL10 concentration correlates with the extent of cellular rejection (P<0.001 by the Jonckheere-Terpstra test). Fetal serum CXCL10 and IL-6 concentrations were shown as median and inter-quartile ranges. *P<0.05; **P<0.01; ***P<0.001 (by the Mann-Whitney U test for comparison between the two groups). ACA, acute chorioamnionitis; CCA, chronic chorioamnionitis; CDP, chronic deciduitis with plasma cells; NS, not significant; VUE, villitis of unknown etiology.
Antibody-mediated Rejection and Fetal Blood CXCL10 Concentration
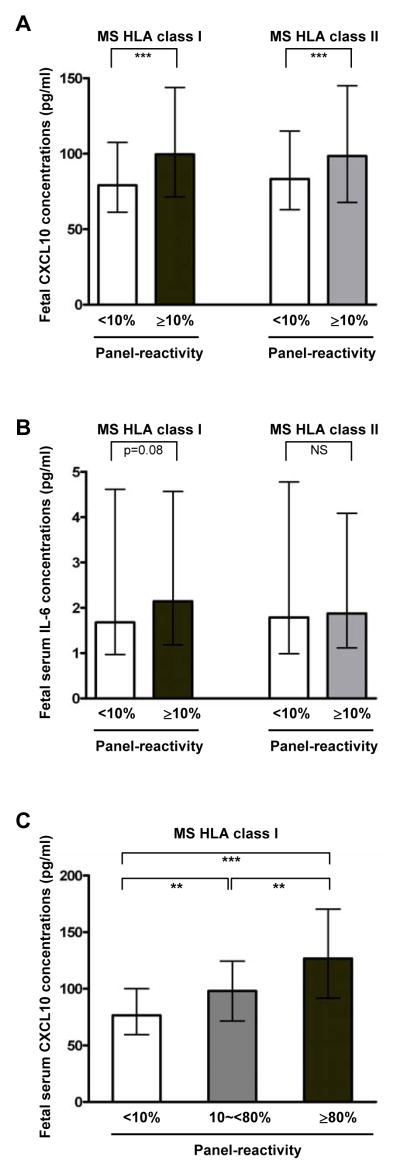
The median fetal serum CXCL10 (but not IL-6) concentration was higher in maternal HLA class I PRA-positive cases than in PRA-negative cases (median 111.5 pg/mL, IQR 80.3–157.2 pg/mL versus median 76.6 pg/mL, IQR 59.5–100.1 pg/mL, P<0.001, Fig. 2A and 2B). A similar difference was also found with HLA class II PRA positivity (median 122.5 pg/mL, IQR 85.5–177.6 pg/mL versus median 81.1 pg/mL, IQR 62.4–113.6 pg/mL, P<0.001). When cases were graded as negative (PRA<10%), mildly sensitized (PRA≥10% and <80%), and highly sensitized (PRA≥80%) according to the reactivity of maternal HLA class I PRA,134,140,141 there was a significant correlation between fetal serum CXCL10 concentration and the degree of maternal sensitization (Fig. 2C). Similar differences in fetal serum CXCL10 concentration were also found in accord with maternal HLA class II PRA positivity.
Figure 2. Fetal serum CXCL10 and IL-6 concentrations according to the presence or absence of maternal HLA PRA.
(A) Median fetal serum CXCL10 concentration is higher in maternal HLA class I PRA-positive cases than in PRA-negative cases (P<0.001). Similar findings were shown between maternal HLA class II PRA-positive and PRA-negative cases. (B) Fetal serum IL-6 concentration was not different according to maternal HLA class I or class II PRA positivity. (C) There was a significant upward trend in fetal serum CXCL10 concentration associated with the degree of maternal HLA sensitization (P<0.001 by the Jonckheere-Terpstra test). Fetal serum CXCL10 and IL-6 concentrations were shown as median and inter-quartile ranges. *P<0.05; **P<0.01; ***P<0.001 (by the Mann-Whitney U test for comparison between the two groups). HLA, human leukocyte antigen; MS, maternal serum; NS, not significant; PRA, panel-reactive antibodies.
Whole-Genome DASL Assay of the Blood Transcriptome
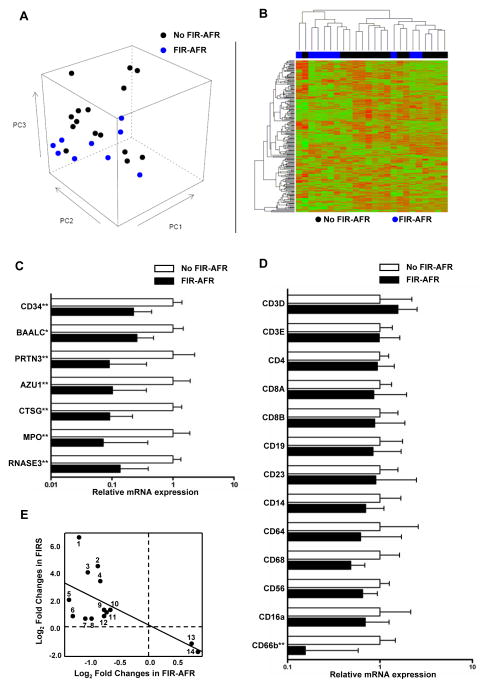
To characterize the blood transcriptome in cases with fetal inflammatory response associated with maternal anti-fetal rejection, Whole-Genome DASL® Assay was performed using fetal blood samples from patients with evidence of fetal inflammatory response associated with maternal anti-fetal rejection (fetal inflammatory response associated with maternal anti-fetal rejection: two or more parameters of cellular rejection, maternal HLA class I PRA ≥80%, and fetal serum CXCL10 concentration >75th percentile) and those without evidence of fetal inflammatory response associated with maternal anti-fetal rejection (no cellular rejection, negative maternal HLA class I and class II PRA, and fetal serum CXCL10 concentration <25th percentile) (Fig. 3A and 3B). A total of 128 genes were differentially expressed in the WBCs of fetuses with and without evidence of fetal inflammatory response associated with maternal anti-fetal rejection (Table II). CD34, BAALC (brain and acute leukemia, cytoplasmic), PRTN3 (proteinase 3), AZU1 (azurocidin 1), CTSG (cathepsin G), MPO (myeloperoxidase), and RNASE3 (ribonuclease, RNase A family, 3) were among the 98 genes whose expression was decreased in cases with evidence of fetal inflammatory response associated with maternal anti-fetal rejection. Differential expression of these genes was confirmed by qRT-PCR along with the decreased mRNA expression of CD66b (but not of CD3, CD4, CD8, CD14, CD16a, CD19, CD23, CD56, CD64, and CD68) in the blood of fetuses with evidence of fetal inflammatory response associated with maternal anti-fetal rejection (Fig. 3C and 3D). Gene Ontology analysis of differentially expressed genes showed enrichment of 24 biological processes such as ‘response to other organism’ and ‘killing by host of symbiont cells’ (Table III).
Figure 3. Transcriptome analysis of fetal blood using whole genome DASL® assay according to the presence or absence of fetal inflammatory response associated with maternal anti-fetal rejection.
(A) An unsupervised Principal Component Analysis based on expression of all genes on the array shows that samples of the group without fetal inflammatory response associated with maternal anti-fetal rejection tend to have higher PC3 and PC1 coordinates than samples of the fetal inflammatory response associated with maternal anti-fetal rejection group. (B) A clustered heat map based on the top 200 most varying genes shows two main clusters: one dominated by samples of the fetal inflammatory response associated with maternal anti-fetal rejection group (left) and one dominated by samples of the group without fetal inflammatory response associated with maternal anti-fetal rejection (right). (C) Quantitative RT-PCR results confirm differential expression of genes of interest: mRNA expression of CD34, BAALC (brain and acute leukemia, cytoplasmic), PRTN3 (proteinase 3), AZU1 (azurocidin 1), CTSG (cathepsin G), MPO (myeloperoxidase), and RNASE3 (ribonuclease, RNase A family, 3) was decreased in cases with fetal inflammatory response associated with maternal anti-fetal rejection (P<0.05, for each). (D) Quantitative RT-PCR of leukocyte marker genes demonstrates that mRNA expression of CD66b (a marker for polymorphonuclear leukocyte) was decreased in the blood of cases with fetal inflammatory response associated with maternal anti-fetal rejection (P<0.01). However, there was no difference in mRNA expression of T cell markers (CD3D, CD3E, CD4, CD8A, and CD8B), B cell markers (CD19 and CD23), monocyte markers (CD14 and CD64), and natural killer cell or macrophage markers (CD56 and CD68). (E) Comparison of differentially expressed genes between fetal inflammatory response syndrome to intra-amniotic infection (FIRS) shown in a previous study by Madsen-Bouterse et al.6 Fetal inflammatory response associated with maternal anti-fetal rejection cases showed only 14 genes common to both conditions – RETN, LCN2, TCN1, RNASE2, CEBPE, FOXM1, CEP55, C12orf59, CAPN3, TP53I3, TYMS, GINS2, ID3, and FCER2 – and all were inversely correlated. Relative mRNA expressions were shown as median and inter-quartile ranges.
*P < 0.05; **P < 0.01; ***P < 0.001 (by the Mann-Whitney U test). FIR-AFR, fetal inflammatory response syndrome associated with maternal anti-fetal rejection; FIRS, fetal inflammatory response syndrome.
Table II.
Top 25 each of up- and down-regulated genes in fetal inflammatory response associated with maternal anti-fetal rejection
| Gene | Fold-change | P value | Direction |
|---|---|---|---|
| GCET2 | 2.02 | 0.0008 | ↑ |
| EFEMP1 | 1.99 | 0.0071 | ↑ |
| TCEA3 | 1.83 | 0.0000 | ↑ |
| FCER2 | 1.79 | 0.0049 | ↑ |
| FCRL5 | 1.75 | 0.0096 | ↑ |
| SCARNA21 | 1.65 | 0.0090 | ↑ |
| TTC39B | 1.63 | 0.0057 | ↑ |
| ID3 | 1.62 | 0.0090 | ↑ |
| PPAPDC1B | 1.62 | 0.0011 | ↑ |
| GBP1 | 1.61 | 0.0019 | ↑ |
| DKK3 | 1.59 | 0.0043 | ↑ |
| ATPBD4 | 1.59 | 0.0033 | ↑ |
| HAPLN3 | 1.59 | 0.0099 | ↑ |
| AXIN2 | 1.59 | 0.0018 | ↑ |
| GBP1 | 1.57 | 0.0027 | ↑ |
| C6orf105 | 1.57 | 0.0023 | ↑ |
| HPCAL4 | 1.57 | 0.0056 | ↑ |
| NUDT9P1 | 1.57 | 0.0034 | ↑ |
| FAM134B | 1.56 | 0.0059 | ↑ |
| ZNF391 | 1.56 | 0.0030 | ↑ |
| GNB5 | 1.55 | 0.0019 | ↑ |
| ZNF667 | 1.54 | 0.0027 | ↑ |
| LOC100129902 | 1.54 | 0.0036 | ↑ |
| P2RY10 | 1.54 | 0.0073 | ↑ |
| SOCS1 | 1.53 | 0.0062 | ↑ |
| PRTN3 | 7.09 | 0.0000 | ↓ |
| AZU1 | 6.71 | 0.0002 | ↓ |
| CTSG | 4.58 | 0.0002 | ↓ |
| MPO | 4.35 | 0.0003 | ↓ |
| MS4A3 | 4.05 | 0.0001 | ↓ |
| RNASE3 | 3.96 | 0.0003 | ↓ |
| DEFA4 | 3.74 | 0.0018 | ↓ |
| TACSTD2 | 3.73 | 0.0021 | ↓ |
| COL17A1 | 3.72 | 0.0008 | ↓ |
| ELANE | 3.58 | 0.0001 | ↓ |
| CEACAM6 | 3.53 | 0.0005 | ↓ |
| TCTEX1D1 | 3.53 | 0.0004 | ↓ |
| LTF | 3.37 | 0.0059 | ↓ |
| CEACAM8 | 3.31 | 0.0007 | ↓ |
| ABCA13 | 2.95 | 0.0067 | ↓ |
| MS4A3 | 2.83 | 0.0005 | ↓ |
| SERPINB10 | 2.78 | 0.0066 | ↓ |
| SLC2A5 | 2.72 | 0.0062 | ↓ |
| BPI | 2.68 | 0.0017 | ↓ |
| CD34 | 2.60 | 0.0007 | ↓ |
| CEBPE | 2.59 | 0.0047 | ↓ |
| MKI67 | 2.54 | 0.0070 | ↓ |
| FIS | 2.53 | 0.0077 | ↓ |
| FOXM1 | 2.51 | 0.0063 | ↓ |
| CKAP2L | 2.45 | 0.0055 | ↓ |
Table III.
Top biological processes enriched in fetal inflammatory response associated with maternal anti-fetal rejection
| Biological process | No. Differentially Expressed Genes/No. Total Genes | P value | False Discovery Rate |
|---|---|---|---|
| Response to bacterium | 10/145 | 0.0000 | 0.0004 |
| Defense response to bacterium | 7/57 | 0.0000 | 0.0004 |
| Killing of cells of another organism | 4/12 | 0.0000 | 0.0011 |
| Defense response to fungus | 3/8 | 0.0000 | 0.0103 |
| M phase | 12/348 | 0.0001 | 0.0103 |
| Nuclear division | 10/245 | 0.0001 | 0.0103 |
| Mitosis | 10/245 | 0.0001 | 0.0103 |
| Response to other organism | 10/251 | 0.0001 | 0.0103 |
| M phase of mitotic cell cycle | 10/252 | 0.0001 | 0.0103 |
| Organelle fission | 10/254 | 0.0001 | 0.0103 |
| DNA replication | 9/210 | 0.0001 | 0.0114 |
| Neutrophil mediated cytotoxicity | 2/3 | 0.0002 | 0.0216 |
| Neutrophil mediated killing of symbiont cell | 2/3 | 0.0002 | 0.0216 |
| Response to fungus | 3/16 | 0.0004 | 0.0304 |
| Cell division | 10/307 | 0.0004 | 0.0321 |
| Disruption by host of symbiont cells | 2/4 | 0.0005 | 0.0329 |
| Killing by host of symbiont cells | 2/4 | 0.0005 | 0.0329 |
| Cell cycle process | 14/574 | 0.0005 | 0.0352 |
| Cell cycle phase | 12/443 | 0.0006 | 0.0352 |
| Response to biotic stimulus | 10/323 | 0.0006 | 0.0352 |
| Cell killing | 4/44 | 0.0006 | 0.0352 |
| Disruption of cells of other organism involved in symbiotic interaction | 2/5 | 0.0008 | 0.0403 |
| Killing of cells in other organism involved in symbiotic interaction | 2/5 | 0.0008 | 0.0403 |
| Mitotic spindle organization | 3/22 | 0.0009 | 0.0469 |
When we compared differentially expressed genes in cases with evidence of fetal inflammatory response associated with maternal anti-fetal rejection (N=128) with those found to be linked with FIRS-associated intra-amniotic infection (N=448),84 only 14 genes (RETN, LCN2, TCN1, RNASE2, CEBPE, FOXM1, CEP55, C12orf59, CAPN3, TP53I3, TYMS, GINS2, ID3, and FCER2) were common to both conditions, but all were inversely correlated, demonstrating a clear difference between these two conditions (Fig. 3E).
2D-DIGE of Fetal Serum
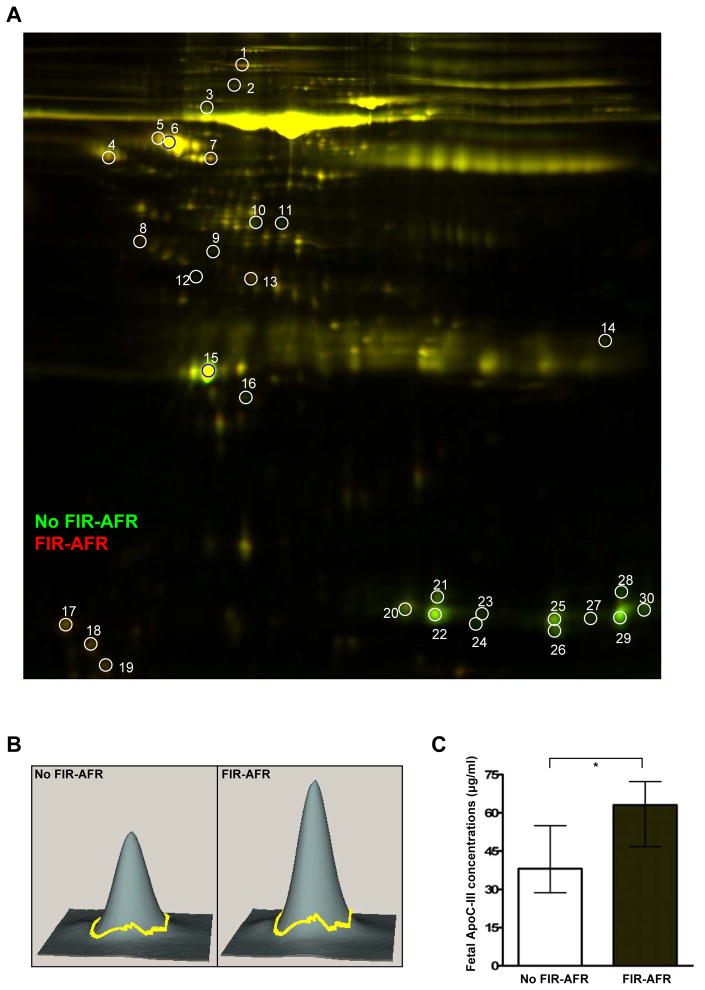
Fig. 4A shows the 2-D electrophoresis gel with 30 spots displaying differentially expressed proteins (more than a 1.5 fold-change) between cases with and without evidence of fetal inflammatory response associated with maternal anti-fetal rejection. Table IV shows the list of 20 differentially expressed proteins identified from 30 spots in the 2D-DIGE analysis of pooled fetal serum samples from each group (cases with and without evidence of fetal inflammatory response associated with maternal anti-fetal rejection). Serum albumin and hemoglobin were decreased in cases with evidence of fetal inflammatory response associated with maternal anti-fetal rejection. Interestingly, several apolipoproteins were found to be differentially abundant between the two groups: apolipoprotein E precursor, apolipoprotein J precursor, and apolipoprotein E3 fragment were decreased, and apolipoprotein C-III was increased, in cases with evidence of fetal inflammatory response associated with maternal anti-fetal rejection.
Figure 4. Comparison of the fetal blood proteome between cases with and without fetal inflammatory response associated with maternal anti-fetal rejection.
(A) Two-dimensional difference gel electrophoresis (2D-DIGE) images show differentially abundant proteins between cases with (Cy5: red) and without (Cy3: green) Fetal inflammatory response associated with maternal anti-fetal rejection. The spots are labeled by number. (B) Three-dimensional images from DeCyder software analysis for spot 17 marked in the 2D-DIGE image (A), which is increased in cases with fetal inflammatory response associated with maternal anti-fetal rejection. The spot was identified as apolipoprotein C-III by MALDI-TOF-MS/MS. (C) The difference in apolipoprotein C-III concentration in fetal serum samples between the cases with and without fetal inflammatory response associated with maternal anti-fetal rejection was confirmed by specific immunoassay (P=0.013).
*P<0.05 (by the Mann-Whitney U test)
ApoC-III, Apolipoprotein C-III; FIR-AFR, fetal inflammatory response syndrome associated with maternal anti-fetal rejection.
Table IV.
Fetal serum proteins show significant changes in cases with fetal inflammatory response associated with maternal anti-fetal rejection
| Spot number | Protein name | Accession number | Molecular weight (Da) | Protein PI |
|---|---|---|---|---|
| 2 | Chain A, Crystal Structure of the Ga Module Complexed with Human Serum Albumin | gi|55669910 | 65178.2 | 5.6 |
| 3 | Alpha-1-B glycoprotein [Homo sapiens] | gi|119592981 | 54238.6 | 5.6 |
| 8 | Apolipoprotein J precursor [Homo sapiens] | gi|178855 | 48772.1 | 6.3 |
| 9 | Apolipoprotein E precursor [Homo sapiens] | gi|4557325 | 36131.8 | 5.7 |
| 11 | Chain B, Crystal Structure Of Fibrinogen Fragment D | gi|2781208 | 37624.7 | 5.8 |
| 12 | Chain A, Apolipoprotein E3 22kd Fragment Lys146gln Mutant | gi|15826034 | 22116.5 | 5.4 |
| 14 | C1q B-chain precursor [Homo sapiens] | gi|573114 | 23925.9 | 8.9 |
| 16 | Peroxiredoxin-2 isoform a [Homo sapiens] | gi|32189392 | 21878.2 | 5.7 |
| 17 | Apolipoprotein C-III [Homo sapiens] | gi|521205 | 10815.5 | 5.2 |
| 20 | Chain G, Structure of Human Foetal Deoxyhaemoglobin | gi|157875419 | 15985.2 | 6.7 |
| 21 | Chain B, Human Hemoglobin A Mutant Beta H63w Carbonmonoxy-Form | gi|300508775 | 15906.3 | 6.8 |
| 22 | Chain G, Structure of Human Foetal Deoxyhaemoglobin | gi|157875419 | 15985.2 | 6.7 |
| 23 | Hemoglobin subunit gamma-2 [Homo sapiens] | gi|6715607 | 16116.3 | 6.6 |
| 24 | Chain A, Solution Structure of Human Normal Adult Hemoglobin | gi|157883730 | 15071.8 | 8.1 |
| 25 | Chain A, Crystal Structure of Oxy-Human Hemoglobin Bassett at 2.15 Angstrom | gi|37928140 | 15072.9 | 9.1 |
| 26 | Hemoglobin alpha-1 globin chain [Homo sapiens] | gi|319739573 | 10776.5 | 8.1 |
| 27 | Chain A, Structure of Haemoglobin in the Deoxy Quaternary State with Ligand Bound at the Alpha Haem | gi|229751 | 15116.9 | 8.7 |
| 28 | Chain A, Structure of Haemoglobin in the Deoxy Quaternary State with Ligand Bound at the Alpha Haem | gi|229751 | 15116.9 | 8.7 |
| 29 | Hemoglobin alpha-1 globin chain [Homo sapiens] | gi|319739573 | 10776.5 | 8.1 |
| 30 | Chain A, Structure of Haemoglobin in the Deoxy Quaternary State with Ligand Bound at the Alpha Haem | gi|229751 | 15116.9 | 8.7 |
To confirm the results of proteins of interest in 2D-DIGE, serum concentrations of apolipoprotein C-III and apolipoprotein E were measured with specific immunoassays. A higher serum concentration of apolipoprotein C-III in cases with evidence of fetal inflammatory response associated with maternal anti-fetal rejection than in those without it was confirmed (fetal inflammatory response associated with maternal anti-fetal rejection: median 63.0 μg/mL, IQR 47.4–69.9 μg/mL versus no evidence of fetal inflammatory response associated with maternal anti-fetal rejection: median 38.1 μg/mL, IQR 29.8–53.4 μg/mL, P=0.013; Fig. 4C), while serum apolipoprotein E concentration was not significantly different between the two groups (evidence of fetal inflammatory response associated with maternal anti-fetal rejection: median 148.6 μg/mL, IQR 137.2–180.2 μg/mL versus no evidence of fetal inflammatory response associated with maternal anti-fetal rejection: median 119.9 μg/mL, IQR 111.6–165.6 μg/mL, P =0.199).
Discussion
Principal findings of this study
1) The frequency of placental lesions consistent with maternal anti-fetal rejection was higher in patients with spontaneous preterm delivery than in those with term delivery; 2) patients with spontaneous preterm births had a higher rate of maternal HLA PRA class I positivity than those who delivered at term; 3) fetuses born of pregnancies with evidence of maternal anti-fetal rejection had a higher fetal serum CXCL10 than those without this process; and 4) the WBC transcriptome and serum proteome were different in those with and without evidence of fetal inflammatory response associated with maternal anti-fetal rejection, suggesting the existence of a distinct form of a systemic inflammatory response in fetuses that were immunologically rejected by their mothers.
The clinical significance of an elevation of CXCL10
CXCL10, a ligand for CXCR3, is chemotactic for activated T cells, macrophages, and NK cells.142–144 Notably, CXCL10 is one of the most commonly expressed chemokines during allograft rejection and GVHD.145–148 An elevated intra-graft CXCL10 expression is associated with renal, lung, and cardiac allograft rejection.149–157 Additionally, an elevated serum CXCL10 concentration before organ transplantation is predictive of poor allograft outcome.151,153,154,158 Our study shows that maternal anti-fetal cellular rejection and antibody-mediated rejection are associated with increased systemic fetal chemokine CXCL10 concentration, as intra-amniotic infection is linked to an elevation of the systemic fetal cytokine IL-6 concentration.31,33 Further, we also demonstrated that maternal anti-fetal rejection shares common features with allograft rejection. Indeed, the current study demonstrates that fetuses with evidence of maternal anti-fetal rejection have elements of an inflammatory response which is quite distinct from that observed in FIRS-associated intra-amniotic infection and acute inflammatory lesions31–33. We proposed the term “fetal inflammatory response syndrome type II” for this condition. The bases for the proposal are that: 1) fetal serum CXCL10 (but not IL-6) concentration is associated with anti-fetal cellular rejection and antibody-mediated rejection, and 2) there are no overlapping changes in the fetal blood transcriptome between fetal inflammatory response associated with maternal anti-fetal rejection and FIRS associated intra-amniotic infection and acute inflammatory lesions, which we will refer to henceforth as “FIRS type I”.
We conducted comprehensive analyses of the fetal blood transcriptome and proteome to characterize fetal systemic changes associated with fetal inflammatory response associated with maternal anti-fetal rejection, and found biologically meaningful changes. BAALC is expressed in CD34+ hematopoietic progenitor cells from bone marrow, and it is a poor prognostic factor in acute myeloid leukemia.159 PRTN3, AZU1, CTSG, MPO, and RNASE3 can also be expressed in CD34+ hematopoietic progenitor cells which are essential for the function of mature neutrophils and eosinophils.160 Universal down-regulation of mRNA expression of the genes for neutrophil granule proteins and the polymorphonuclear leukocyte surface marker (CD66b) is consistent with earlier observations of neonatal alloimmune neutropenia induced by maternal HLA antibodies.161,162 Overall changes in the fetal blood transcriptome strongly suggested the presence of an alloimmune reaction in the fetus probably caused by the deleterious effect of maternal anti-HLA antibodies which cross the placenta and activate complement in the endothelium of the umbilical cord vein.
Among changes in the serum proteins, we found an overexpression of apolipoprotein C-III which was confirmed by immunoassay. Apolipoprotein C-III in plasma has been shown to be associated with coronary heart disease, atherosclerosis, and metabolic syndromes such as obesity, hypertriglyceridemia, and type 2 diabetes.163,164 Therefore, the increase in serum apolipoprotein C-III in fetuses with evidence of fetal inflammatory response associated with maternal anti-fetal rejection is intriguing, and raises the need for further studies about potential long-term consequences of prenatal exposure to maternal immunological rejection. Several studies have clearly indicated that an abnormal intrauterine environment can affect lifelong fetal well-being in the form of abnormal fetal programming.165–167 Individuals exposed to the Dutch famine during pregnancy had a higher frequency of coronary heart disease,168 and also display an atherogenic blood lipid profile as a consequence of metabolic stress in utero.169 Although fetal inflammatory response associated with maternal anti-fetal rejection does not have a direct relationship to maternal nutritional intake, changes in blood lipid profiles strongly suggest that fetal inflammatory response associated with maternal anti-fetal rejection could alter fetal programming.
In organ transplantation, humoral antibody-mediated allograft rejection occurs by different mechanisms from T-cell-mediated rejection.170,171 However, both cell-mediated and antibody-mediated rejections begin with recognition as a common starting point and are followed by: 1) CD4+ and CD8+ T cell cytotoxicity, 2) CD4+ and CD8+ derived IFN-γ production and delayed-type hypersensitivity, and 3) complement activation or antibody-dependent cell-mediated cytotoxicity by antibodies reactive to donor MHC molecules;171 and these three phenomena are closely related to each other. Our previous studies demonstrated a robust association between anti-fetal cellular rejection (chronic chorioamnionitis) and anti-fetal antibody-mediated rejection (both positive maternal HLA PRA and C4d deposition on umbilical vein endothelium),27 and fetal HLA specificity of maternal HLA antibodies.29 The findings in this study also support the hypothesis that anti-fetal antibody-mediated rejection has biological consequences similar to cellular rejection, sharing the feature of an increased CXCL10 concentration in fetal sera.
Strengths and limitations
We described a novel form of fetal systemic inflammation in the context of maternal anti-fetal rejection. The limitations of this study include: 1) we did not define the cut-off value of fetal serum CXCL10 concentration to predict evidence of fetal inflammatory response associated with maternal anti-fetal rejection. This has been defined as FIRS type I with a fetal plasma IL6 concentration above 11 pg/ml in blood obtained by cordocentesis.31,33,48 Future studies are required to identify a cut-off value and also the short- and long-term consequences of this inflammatory process; and 2) the leukocyte counts of cord blood at the time of delivery were not analyzed. As retrospective analysis of cord blood is impossible, we instead compared mRNA expression levels of genes encoding cell-type-specific surface markers of leukocytes.
Conclusions
Collectively, the findings reported herein link maternal anti-fetal rejection with a systemic inflammatory response in the fetus. This inflammatory response is characterized with changes in the fetal blood transcriptome and proteome which are different from FIRS type I associated with acute inflammatory lesions of the placenta. Future studies are required to define pragmatic diagnostic criteria as well as short- and long-term consequences of this condition.
Supplementary Material
Acknowledgments
This research was supported, in part, by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services (NICHD/NIH); and, in part, with Federal funds from NICHD, NIH under Contract No. HSN275201300006C. The authors are grateful to the patients who agreed to participate in our studies; to the nurses, laboratory staff, and clinicians who made this work possible; and to Maureen McGerty and Andrea Bernard (Wayne State University) for their critical readings of the manuscript.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest.
References
- 1.Mor G, Cardenas I. The immune system in pregnancy: a unique complexity. Am J Reprod Immunol. 2010;63:425–433. doi: 10.1111/j.1600-0897.2010.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mor G, Cardenas I, Abrahams V, Guller S. Inflammation and pregnancy: the role of the immune system at the implantation site. Ann N Y Acad Sci. 2011;1221:80–87. doi: 10.1111/j.1749-6632.2010.05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotlieb WH. Immunology of pregnancy. Rev Med Brux. 1992;13:97–101. [PubMed] [Google Scholar]
- 4.Terness P, Kallikourdis M, Betz AG, Rabinovich GA, Saito S, Clark DA. Tolerance signaling molecules and pregnancy: IDO, galectins, and the renaissance of regulatory T cells. Am J Reprod Immunol. 2007;58:238–254. doi: 10.1111/j.1600-0897.2007.00510.x. [DOI] [PubMed] [Google Scholar]
- 5.Blois SM, Ilarregui JM, Tometten M, Garcia M, Orsal AS, Cordo-Russo R, Toscano MA, Bianco GA, Kobelt P, Handjiski B, Tirado I, Markert UR, Klapp BF, Poirier F, Szekeres-Bartho J, Rabinovich GA, Arck PC. A pivotal role for galectin-1 in fetomaternal tolerance. Nat Med. 2007;13:1450–1457. doi: 10.1038/nm1680. [DOI] [PubMed] [Google Scholar]
- 6.Than NG, Romero R, Erez O, Weckle A, Tarca AL, Hotra J, Abbas A, Han YM, Kim SS, Kusanovic JP, Gotsch F, Hou Z, Santolaya-Forgas J, Benirschke K, Papp Z, Grossman LI, Goodman M, Wildman DE. Emergence of hormonal and redox regulation of galectin-1 in placental mammals: implication in maternal-fetal immune tolerance. Proc Natl Acad Sci U S A. 2008;105:15819–15824. doi: 10.1073/pnas.0807606105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Than NG, Romero R, Goodman M, Weckle A, Xing J, Dong Z, Xu Y, Tarquini F, Szilagyi A, Gal P, Hou Z, Tarca AL, Kim CJ, Kim JS, Haidarian S, Uddin M, Bohn H, Benirschke K, Santolaya-Forgas J, Grossman LI, Erez O, Hassan SS, Zavodszky P, Papp Z, Wildman DE. A primate subfamily of galectins expressed at the maternal-fetal interface that promote immune cell death. Proc Natl Acad Sci U S A. 2009;106:9731–9736. doi: 10.1073/pnas.0903568106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Than NG, Romero R, Kim CJ, McGowen MR, Papp Z, Wildman DE. Galectins: guardians of eutherian pregnancy at the maternal-fetal interface. Trends Endocrinol Metab. 2012;23:23–31. doi: 10.1016/j.tem.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mellor AL, Munn DH. Tryptophan catabolism prevents maternal T cells from activating lethal anti-fetal immune responses. J Reprod Immunol. 2001;52:5–13. doi: 10.1016/s0165-0378(01)00118-8. [DOI] [PubMed] [Google Scholar]
- 10.Chaouat G, Voisin GA, Daeron M, Kanellopoulos J. Enhancing antibodies and supressive cells in maternal anti-fetal immune reaction. Ann Immunol (Paris) 1977;128:21–24. [PubMed] [Google Scholar]
- 11.Trowsdale J, Betz AG. Mother’s little helpers: mechanisms of maternal-fetal tolerance. Nat Immunol. 2006;7:241–246. doi: 10.1038/ni1317. [DOI] [PubMed] [Google Scholar]
- 12.Abrahams VM, Straszewski-Chavez SL, Guller S, Mor G. First trimester trophoblast cells secrete Fas ligand which induces immune cell apoptosis. Mol Hum Reprod. 2004;10:55–63. doi: 10.1093/molehr/gah006. [DOI] [PubMed] [Google Scholar]
- 13.Thaxton JE, Sharma S. Interleukin-10: a multi-faceted agent of pregnancy. Am J Reprod Immunol. 2010;63:482–491. doi: 10.1111/j.1600-0897.2010.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mor G. Inflammation and pregnancy: the role of toll-like receptors in trophoblast-immune interaction. Ann N Y Acad Sci. 2008;1127:121–128. doi: 10.1196/annals.1434.006. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Koga K, Osuga Y, Cardenas I, Izumi G, Takamura M, Hirata T, Yoshino O, Hirota Y, Harada M, Mor G, Taketani Y. Toll-like receptor-3 ligation-induced indoleamine 2, 3-dioxygenase expression in human trophoblasts. Endocrinology. 2011;152:4984–4992. doi: 10.1210/en.2011-0278. [DOI] [PubMed] [Google Scholar]
- 16.Kalkunte S, Chichester CO, Gotsch F, Sentman CL, Romero R, Sharma S. Evolution of non-cytotoxic uterine natural killer cells. Am J Reprod Immunol. 2008;59:425–432. doi: 10.1111/j.1600-0897.2008.00595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Than NG, Nagai A, Sharma S, Meiri H, Hauguel-de Mouzon S, Sadovsky Y, Rao Ch V. Application of pregnancy-related proteins in prenatal and tumor diagnostics--a workshop report. Placenta. 2005;26 (Suppl A):S110–113. doi: 10.1016/j.placenta.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Schober L, Radnai D, Schmitt E, Mahnke K, Sohn C, Steinborn A. Term and preterm labor: decreased suppressive activity and changes in composition of the regulatory T-cell pool. Immunol Cell Biol. 2012;90:935–944. doi: 10.1038/icb.2012.33. [DOI] [PubMed] [Google Scholar]
- 19.Steinborn A, Schmitt E, Kisielewicz A, Rechenberg S, Seissler N, Mahnke K, Schaier M, Zeier M, Sohn C. Pregnancy-associated diseases are characterized by the composition of the systemic regulatory T cell (Treg) pool with distinct subsets of Tregs. Clin Exp Immunol. 2012;167:84–98. doi: 10.1111/j.1365-2249.2011.04493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito S, Sakai M, Sasaki Y, Nakashima A, Shiozaki A. Inadequate tolerance induction may induce pre-eclampsia. J Reprod Immunol. 2007;76:30–39. doi: 10.1016/j.jri.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki Y, Darmochwal-Kolarz D, Suzuki D, Sakai M, Ito M, Shima T, Shiozaki A, Rolinski J, Saito S. Proportion of peripheral blood and decidual CD4(+) CD25(bright) regulatory T cells in pre-eclampsia. Clin Exp Immunol. 2007;149:139–145. doi: 10.1111/j.1365-2249.2007.03397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tirado-Gonzalez I, Freitag N, Barrientos G, Shaikly V, Nagaeva O, Strand M, Kjellberg L, Klapp BF, Mincheva-Nilsson L, Cohen M, Blois SM. Galectin-1 influences trophoblast immune evasion and emerges as a predictive factor for the outcome of pregnancy. Mol Hum Reprod. 2012;19:43–53. doi: 10.1093/molehr/gas043. [DOI] [PubMed] [Google Scholar]
- 23.Kisielewicz A, Schaier M, Schmitt E, Hug F, Haensch GM, Meuer S, Zeier M, Sohn C, Steinborn A. A distinct subset of HLA-DR+-regulatory T cells is involved in the induction of preterm labor during pregnancy and in the induction of organ rejection after transplantation. Clin Immunol. 2010;137:209–220. doi: 10.1016/j.clim.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Steinborn A, Saran G, Schneider A, Fersis N, Sohn C, Schmitt E. The presence of gestational diabetes is associated with increased detection of anti-HLA-class II antibodies in the maternal circulation. Am J Reprod Immunol. 2006;56:124–134. doi: 10.1111/j.1600-0897.2006.00408.x. [DOI] [PubMed] [Google Scholar]
- 25.Steinborn A, Schmitt E, Stein Y, Klee A, Gonser M, Seifried E, Seidl C. Prolonged preterm rupture of fetal membranes, a consequence of an increased maternal anti-fetal T cell responsiveness. Pediatr Res. 2005;58:648–653. doi: 10.1203/01.PDR.0000180541.03425.76. [DOI] [PubMed] [Google Scholar]
- 26.Kim CJ, Romero R, Kusanovic JP, Yoo W, Dong Z, Topping V, Gotsch F, Yoon BH, Chi JG, Kim JS. The frequency, clinical significance, and pathological features of chronic chorioamnionitis: a lesion associated with spontaneous preterm birth. Mod Pathol. 2010;23:1000–1011. doi: 10.1038/modpathol.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Romero R, Xu Y, Kim JS, Topping V, Yoo W, Kusanovic JP, Chaiworapongsa T, Hassan SS, Yoon BH, Kim CJ. A signature of maternal anti-fetal rejection in spontaneous preterm birth: chronic chorioamnionitis, anti-human leukocyte antigen antibodies, and C4d. PLoS One. 2011;6:e16806. doi: 10.1371/journal.pone.0016806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Romero R, Dong Z, Xu Y, Qureshi F, Jacques S, Yoo W, Chaiworapongsa T, Mittal P, Hassan SS, Kim CJ. Unexplained fetal death has a biological signature of maternal anti-fetal rejection: chronic chorioamnionitis and alloimmune anti-human leucocyte antigen antibodies. Histopathology. 2011;59:928–938. doi: 10.1111/j.1365-2559.2011.04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J, Romero R, Xu Y, Kim JS, Park JY, Kusanovic JP, Chaiworapongsa T, Hassan SS, Kim CJ. Maternal HLA panel-reactive antibodies in early gestation positively correlate with chronic chorioamnionitis: evidence in support of the chronic nature of maternal anti-fetal rejection. Am J Reprod Immunol. 2011;66:510–526. doi: 10.1111/j.1600-0897.2011.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim MJ, Romero R, Kim CJ, Tarca AL, Chhauy S, LaJeunesse C, Lee DC, Draghici S, Gotsch F, Kusanovic JP, Hassan SS, Kim JS. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. J Immunol. 2009;182:3919–3927. doi: 10.4049/jimmunol.0803834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. The fetal inflammatory response syndrome. Am J Obstet Gynecol. 1998;179:194–202. doi: 10.1016/s0002-9378(98)70272-8. [DOI] [PubMed] [Google Scholar]
- 32.Gotsch F, Romero R, Kusanovic JP, Mazaki-Tovi S, Pineles BL, Erez O, Espinoza J, Hassan SS. The fetal inflammatory response syndrome. Clin Obstet Gynecol. 2007;50:652–683. doi: 10.1097/GRF.0b013e31811ebef6. [DOI] [PubMed] [Google Scholar]
- 33.Romero R, Gomez R, Ghezzi F, Yoon BH, Mazor M, Edwin SS, Berry SM. A fetal systemic inflammatory response is followed by the spontaneous onset of preterm parturition. Am J Obstet Gynecol. 1998;179:186–193. doi: 10.1016/s0002-9378(98)70271-6. [DOI] [PubMed] [Google Scholar]
- 34.Goepfert AR, Andrews WW, Carlo W, Ramsey PS, Cliver SP, Goldenberg RL, Hauth JC. Umbilical cord plasma interleukin-6 concentrations in preterm infants and risk of neonatal morbidity. Am J Obstet Gynecol. 2004;191:1375–1381. doi: 10.1016/j.ajog.2004.06.086. [DOI] [PubMed] [Google Scholar]
- 35.Chaiworapongsa T, Romero R, Kim JC, Kim YM, Blackwell SC, Yoon BH, Gomez R. Evidence for fetal involvement in the pathologic process of clinical chorioamnionitis. Am J Obstet Gynecol. 2002;186:1178–1182. doi: 10.1067/mob.2002.124042. [DOI] [PubMed] [Google Scholar]
- 36.Romero R, Chaiworapongsa T, Espinoza J. Micronutrients and intrauterine infection, preterm birth and the fetal inflammatory response syndrome. J Nutr. 2003;133:1668S–1673S. doi: 10.1093/jn/133.5.1668S. [DOI] [PubMed] [Google Scholar]
- 37.Lee SE, Romero R, Kim CJ, Shim SS, Yoon BH. Funisitis in term pregnancy is associated with microbial invasion of the amniotic cavity and intra-amniotic inflammation. J Matern Fetal Neonatal Med. 2006;19:693–697. doi: 10.1080/14767050600927353. [DOI] [PubMed] [Google Scholar]
- 38.Lee SE, Romero R, Lee SM, Yoon BH. Amniotic fluid volume in intra-amniotic inflammation with and without culture-proven amniotic fluid infection in preterm premature rupture of membranes. J Perinat Med. 2010;38:39–44. doi: 10.1515/JPM.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gervasi MT, Romero R, Bracalente G, Chaiworapongsa T, Erez O, Dong Z, Hassan SS, Yeo L, Yoon BH, Mor G, Barzon L, Franchin E, Militello V, Palu G. Viral invasion of the amniotic cavity (VIAC) in the midtrimester of pregnancy. J Matern Fetal Neonatal Med. 2012;25:2002–2013. doi: 10.3109/14767058.2012.683899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittal P, Romero R, Kusanovic JP, Edwin SS, Gotsch F, Mazaki-Tovi S, Espinoza J, Erez O, Nhan-Chang CL, Than NG, Vaisbuch E, Hassan SS. CXCL6 (granulocyte chemotactic protein-2): a novel chemokine involved in the innate immune response of the amniotic cavity. Am J Reprod Immunol. 2008;60:246–257. doi: 10.1111/j.1600-0897.2008.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotsch F, Romero R, Kusanovic JP, Erez O, Espinoza J, Kim CJ, Vaisbuch E, Than NG, Mazaki-Tovi S, Chaiworapongsa T, Mazor M, Yoon BH, Edwin S, Gomez R, Mittal P, Hassan SS, Sharma S. The anti-inflammatory limb of the immune response in preterm labor, intra-amniotic infection/inflammation, and spontaneous parturition at term: a role for interleukin-10. J Matern Fetal Neonatal Med. 2008;21:529–547. doi: 10.1080/14767050802127349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romero R, Maymon E, Pacora P, Gomez R, Mazor M, Yoon BH, Berry SM. Further observations on the fetal inflammatory response syndrome: a potential homeostatic role for the soluble receptors of tumor necrosis factor alpha. Am J Obstet Gynecol. 2000;183:1070–1077. doi: 10.1067/mob.2000.108885. [DOI] [PubMed] [Google Scholar]
- 43.Romero R, Savasan ZA, Chaiworapongsa T, Berry SM, Kusanovic JP, Hassan SS, Yoon BH, Edwin S, Mazor M. Hematologic profile of the fetus with systemic inflammatory response syndrome. J Perinat Med. 2011;40:19–32. doi: 10.1515/JPM.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaiworapongsa T, Romero R, Berry SM, Hassan SS, Yoon BH, Edwin S, Mazor M. The role of granulocyte colony-stimulating factor in the neutrophilia observed in the fetal inflammatory response syndrome. J Perinat Med. 2011;39:653–666. doi: 10.1515/JPM.2011.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuksman RB, Mazzitelli NG. Second-trimester histopathological placental findings in maternal-fetal inflammatory response syndrome. Pediatr Dev Pathol. 2009;12:42–46. doi: 10.2350/07-09-0354.1. [DOI] [PubMed] [Google Scholar]
- 46.Arad I, Ergaz Z. The fetal inflammatory response syndrome and associated infant morbidity. Isr Med Assoc J. 2004;6:766–769. [PubMed] [Google Scholar]
- 47.Erdei G, Toth P, Vasarhelyi B. New clinical entity in perinatology: fetal inflammatory response syndrome. Orv Hetil. 2003;144:1515–1519. [PubMed] [Google Scholar]
- 48.Pacora P, Chaiworapongsa T, Maymon E, Kim YM, Gomez R, Yoon BH, Ghezzi F, Berry SM, Qureshi F, Jacques SM, Kim JC, Kadar N, Romero R. Funisitis and chorionic vasculitis: the histological counterpart of the fetal inflammatory response syndrome. J Matern Fetal Neonatal Med. 2002;11:18–25. doi: 10.1080/jmf.11.1.18.25. [DOI] [PubMed] [Google Scholar]
- 49.Park JS, Romero R, Yoon BH, Moon JB, Oh SY, Han SY, Ko EM. The relationship between amniotic fluid matrix metalloproteinase-8 and funisitis. Am J Obstet Gynecol. 2001;185:1156–1161. doi: 10.1067/mob.2001.117679. [DOI] [PubMed] [Google Scholar]
- 50.Gravett MG, Rubens CE, Nunes TM. Global report on preterm birth and stillbirth (2 of 7): discovery science. BMC Pregnancy Childbirth. 2010;10 (Suppl 1):S2. doi: 10.1186/1471-2393-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gantert M, Been JV, Gavilanes AW, Garnier Y, Zimmermann LJ, Kramer BW. Chorioamnionitis: a multiorgan disease of the fetus? J Perinatol. 2010;30 (Suppl):S21–30. doi: 10.1038/jp.2010.96. [DOI] [PubMed] [Google Scholar]
- 52.Sergeeva VA, Nesterenko SN, Shabalov NP, Aleksandrovich Iu S. Fetal inflammatory response in the development of multiple organ dysfunction in newborn. Anesteziol Reanimatol. 2010:30–34. [PubMed] [Google Scholar]
- 53.Kim YM, Romero R, Chaiworapongsa T, Espinoza J, Mor G, Kim CJ. Dermatitis as a component of the fetal inflammatory response syndrome is associated with activation of Toll-like receptors in epidermal keratinocytes. Histopathology. 2006;49:506–514. doi: 10.1111/j.1365-2559.2006.02542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Been JV, Lievense S, Zimmermann LJ, Kramer BW, Wolfs TG. Chorioamnionitis as a Risk Factor for Necrotizing Enterocolitis: A Systematic Review and Meta-Analysis. J Pediatr. 2012 doi: 10.1016/j.jpeds.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 55.Athayde N, Wang J, Wang X, Trudinger B. Fetuses delivered following preterm prelabor rupture of the membranes are capable of stimulating a proinflammatory response in endothelial cells. J Soc Gynecol Investig. 2005;12:118–122. doi: 10.1016/j.jsgi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Morais Pereira LH, Pacheco Olegario JG, Rocha LP, de Oliveira Guimaraes CS, Ramalho FS, Dos Reis MA, Miranda Correa RR. Association Between the Markers of FIRS and the Morphologic Alterations in the Liver of Neonates Autopsied in the Perinatal Period. Fetal Pediatr Pathol. 2013;31:48–54. doi: 10.3109/15513815.2012.659536. [DOI] [PubMed] [Google Scholar]
- 57.Sood BG, Madan A, Saha S, Schendel D, Thorsen P, Skogstrand K, Hougaard D, Shankaran S, Carlo W. Perinatal systemic inflammatory response syndrome and retinopathy of prematurity. Pediatr Res. 2010;67:394–400. doi: 10.1203/PDR.0b013e3181d01a36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romero R, Soto E, Berry SM, Hassan SS, Kusanovic JP, Yoon BH, Edwin S, Mazor M, Chaiworapongsa T. Blood pH and gases in fetuses in preterm labor with and without systemic inflammatory response syndrome. J Matern Fetal Neonatal Med. 2012;25:1160–1170. doi: 10.3109/14767058.2011.629247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pereira L, Reddy AP, Alexander AL, Lu X, Lapidus JA, Gravett MG, Nagalla SR. Insights into the multifactorial nature of preterm birth: proteomic profiling of the maternal serum glycoproteome and maternal serum peptidome among women in preterm labor. Am J Obstet Gynecol. 2010;202:555 e551–510. doi: 10.1016/j.ajog.2010.02.048. [DOI] [PubMed] [Google Scholar]
- 60.De Felice C, Bagnoli F, Toti P, Musaro MA, Peruzzi L, Paffetti P, Latini G. Transient hypothyroxinemia of prematurity and histological chorioamnionitis. J Perinat Med. 2005;33:514–518. doi: 10.1515/JPM.2005.091. [DOI] [PubMed] [Google Scholar]
- 61.Lucovnik M, Kornhauser-Cerar L, Premru-Srsen T, Gmeiner-Stopar T, Derganc M. Neutrophil defensins but not interleukin-6 in vaginal fluid after preterm premature rupture of membranes predict fetal/neonatal inflammation and infant neurological impairment. Acta Obstet Gynecol Scand. 2011;90:908–916. doi: 10.1111/j.1600-0412.2011.01177.x. [DOI] [PubMed] [Google Scholar]
- 62.Kim SK, Romero R, Chaiworapongsa T, Kusanovic JP, Mazaki-Tovi S, Mittal P, Erez O, Vaisbuch E, Gotsch F, Pacora P, Yeo L, Gervasi MT, Lamont RF, Yoon BH, Hassan SS. Evidence of changes in the immunophenotype and metabolic characteristics (intracellular reactive oxygen radicals) of fetal, but not maternal, monocytes and granulocytes in the fetal inflammatory response syndrome. J Perinat Med. 2009;37:543–552. doi: 10.1515/JPM.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.El-Haieg DO, Zidan AA, El-Nemr MM. The relationship between sonographic fetal thymus size and the components of the systemic fetal inflammatory response syndrome in women with preterm prelabour rupture of membranes. BJOG. 2008;115:836–841. doi: 10.1111/j.1471-0528.2008.01715.x. [DOI] [PubMed] [Google Scholar]
- 64.Di Naro E, Cromi A, Ghezzi F, Raio L, Uccella S, D’Addario V, Loverro G. Fetal thymic involution: a sonographic marker of the fetal inflammatory response syndrome. Am J Obstet Gynecol. 2006;194:153–159. doi: 10.1016/j.ajog.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 65.Toti P, De Felice C, Stumpo M, Schurfeld K, Di Leo L, Vatti R, Bianciardi G, Buonocore G, Seemayer TA, Luzi P. Acute thymic involution in fetuses and neonates with chorioamnionitis. Hum Pathol. 2000;31:1121–1128. doi: 10.1053/hupa.2000.16676. [DOI] [PubMed] [Google Scholar]
- 66.Wolfs TG, Jellema RK, Turrisi G, Becucci E, Buonocore G, Kramer BW. Inflammation-induced immune suppression of the fetus: a potential link between chorioamnionitis and postnatal early onset sepsis. J Matern Fetal Neonatal Med. 2012;25 (Suppl 1):8–11. doi: 10.3109/14767058.2012.664447. [DOI] [PubMed] [Google Scholar]
- 67.Speer CP. Neonatal respiratory distress syndrome: an inflammatory disease? Neonatology. 2011;99:316–319. doi: 10.1159/000326619. [DOI] [PubMed] [Google Scholar]
- 68.Lee J, Oh KJ, Park CW, Park JS, Jun JK, Yoon BH. The presence of funisitis is associated with a decreased risk for the development of neonatal respiratory distress syndrome. Placenta. 2011;32:235–240. doi: 10.1016/j.placenta.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 69.Lahra MM, Beeby PJ, Jeffery HE. Intrauterine inflammation, neonatal sepsis, and chronic lung disease: a 13-year hospital cohort study. Pediatrics. 2009;123:1314–1319. doi: 10.1542/peds.2008-0656. [DOI] [PubMed] [Google Scholar]
- 70.Kramer BW. Antenatal inflammation and lung injury: prenatal origin of neonatal disease. J Perinatol. 2008;28 (Suppl 1):S21–27. doi: 10.1038/jp.2008.46. [DOI] [PubMed] [Google Scholar]
- 71.Mittendorf R, Covert R, Montag AG, elMasri W, Muraskas J, Lee KS, Pryde PG. Special relationships between fetal inflammatory response syndrome and bronchopulmonary dysplasia in neonates. J Perinat Med. 2005;33:428–434. doi: 10.1515/JPM.2005.076. [DOI] [PubMed] [Google Scholar]
- 72.Sweet DG, Halliday HL. Modeling and remodeling of the lung in neonatal chronic lung disease: implications for therapy. Treat Respir Med. 2005;4:347–359. doi: 10.2165/00151829-200504050-00006. [DOI] [PubMed] [Google Scholar]
- 73.Blanco-Quiros A, Arranz E, Solis G, Garrote JA, Mayo A. High cord blood IL-10 levels in preterm newborns with respiratory distress syndrome. Allergol Immunopathol (Madr) 2004;32:189–196. doi: 10.1016/s0301-0546(04)79238-1. [DOI] [PubMed] [Google Scholar]
- 74.Hallman M. Cytokines, pulmonary surfactant and consequences of intrauterine infection. Biol Neonate. 1999;76 (Suppl 1):2–9. doi: 10.1159/000047039. [DOI] [PubMed] [Google Scholar]
- 75.Nishimaki S, Sato M, An H, Shima Y, Akaike T, Yokoyama U, Yokota S. Comparison of markers for fetal inflammatory response syndrome: fetal blood interleukin-6 and neonatal urinary beta(2)-microglobulin. J Obstet Gynaecol Res. 2009;35:472–476. doi: 10.1111/j.1447-0756.2008.00988.x. [DOI] [PubMed] [Google Scholar]
- 76.Letti Muller AL, de Barrios PM, Kliemann LM, Valerio EG, Gasnier R, Magalhaes JA. Tei index to assess fetal cardiac performance in fetuses at risk for fetal inflammatory response syndrome. Ultrasound Obstet Gynecol. 2010;36:26–31. doi: 10.1002/uog.7584. [DOI] [PubMed] [Google Scholar]
- 77.Romero R, Espinoza J, Goncalves LF, Gomez R, Medina L, Silva M, Chaiworapongsa T, Yoon BH, Ghezzi F, Lee W, Treadwell M, Berry SM, Maymon E, Mazor M, DeVore G. Fetal cardiac dysfunction in preterm premature rupture of membranes. J Matern Fetal Neonatal Med. 2004;16:146–157. doi: 10.1080/14767050400009279. [DOI] [PubMed] [Google Scholar]
- 78.Kuypers E, Ophelders D, Jellema RK, Kunzmann S, Gavilanes AW, Kramer BW. White matter injury following fetal inflammatory response syndrome induced by chorioamnionitis and fetal sepsis: Lessons from experimental ovine models. Early Hum Dev. 2012;88:931–936. doi: 10.1016/j.earlhumdev.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 79.Resch B, Radinger A, Mannhalter C, Binder A, Haas J, Muller WD. Interleukin-6 G(--174)C polymorphism is associated with mental retardation in cystic periventricular leucomalacia in preterm infants. Arch Dis Child Fetal Neonatal Ed. 2009;94:F304–306. doi: 10.1136/adc.2008.140244. [DOI] [PubMed] [Google Scholar]
- 80.Bashiri A, Burstein E, Mazor M. Cerebral palsy and fetal inflammatory response syndrome: a review. J Perinat Med. 2006;34:5–12. doi: 10.1515/JPM.2006.001. [DOI] [PubMed] [Google Scholar]
- 81.Mittendorf R, Montag AG, MacMillan W, Janeczek S, Pryde PG, Besinger RE, Gianopoulos JG, Roizen N. Components of the systemic fetal inflammatory response syndrome as predictors of impaired neurologic outcomes in children. Am J Obstet Gynecol. 2003;188:1438–1434. doi: 10.1067/mob.2003.380. discussion 1444–1436. [DOI] [PubMed] [Google Scholar]
- 82.Svigos JM. The fetal inflammatory response syndrome and cerebral palsy: yet another challenge and dilemma for the obstetrician. Aust N Z J Obstet Gynaecol. 2001;41:170–176. doi: 10.1111/j.1479-828x.2001.tb01203.x. [DOI] [PubMed] [Google Scholar]
- 83.Lautridou A, Ancel PY, Launay E, Denizot S, Orsonneau JL, Roze JC, Gras-Le Guen C. Umbilical cord blood procalcitonin as a risk factor for mortality in very premature infants. Eur J Clin Microbiol Infect Dis. 2012;31:2407–2412. doi: 10.1007/s10096-012-1583-0. [DOI] [PubMed] [Google Scholar]
- 84.Madsen-Bouterse SA, Romero R, Tarca AL, Kusanovic JP, Espinoza J, Kim CJ, Kim JS, Edwin SS, Gomez R, Draghici S. The transcriptome of the fetal inflammatory response syndrome. Am J Reprod Immunol. 2010;63:73–92. doi: 10.1111/j.1600-0897.2009.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kanamori Y, Terawaki K, Takayasu H, Sugiyama M, Komura M, Kodaka T, Suzuki K, Kitano Y, Kuroda T, Iwanaka T. Interleukin 6 and interleukin 8 play important roles in systemic inflammatory response syndrome of meconium peritonitis. Surg Today. 2012;42:431–434. doi: 10.1007/s00595-011-0034-3. [DOI] [PubMed] [Google Scholar]
- 86.Savasan ZA, Chaiworapongsa T, Romero R, Hussein Y, Kusanovic JP, Xu Y, Dong Z, Kim CJ, Hassan SS. Interleukin-19 in fetal systemic inflammation. J Matern Fetal Neonatal Med. 2012;25:995–1005. doi: 10.3109/14767058.2011.605917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ozdemir O, Dinleyici EC, Tekin N, Colak O, Aksit MA. Low-mannose-binding lectin levels in susceptibility to neonatal sepsis in preterm neonates with fetal inflammatory response syndrome. J Matern Fetal Neonatal Med. 2010;23:1009–1013. doi: 10.3109/14767050903551418. [DOI] [PubMed] [Google Scholar]
- 88.Salvesen B, Fung M, Saugstad OD, Mollnes TE. Role of complement and CD14 in meconium-induced cytokine formation. Pediatrics. 2008;121:e496–505. doi: 10.1542/peds.2007-0878. [DOI] [PubMed] [Google Scholar]
- 89.Yoon BH, Romero R, Park JS, Kim M, Oh SY, Kim CJ, Jun JK. The relationship among inflammatory lesions of the umbilical cord (funisitis), umbilical cord plasma interleukin 6 concentration, amniotic fluid infection, and neonatal sepsis. Am J Obstet Gynecol. 2000;183:1124–1129. doi: 10.1067/mob.2000.109035. [DOI] [PubMed] [Google Scholar]
- 90.Sadowsky DW, Adams KM, Gravett MG, Witkin SS, Novy MJ. Preterm labor is induced by intraamniotic infusions of interleukin-1beta and tumor necrosis factor-alpha but not by interleukin-6 or interleukin-8 in a nonhuman primate model. Am J Obstet Gynecol. 2006;195:1578–1589. doi: 10.1016/j.ajog.2006.06.072. [DOI] [PubMed] [Google Scholar]
- 91.Collins JJ, Kallapur SG, Knox CL, Nitsos I, Polglase GR, Pillow JJ, Kuypers E, Newnham JP, Jobe AH, Kramer BW. Inflammation in fetal sheep from intra-amniotic injection of Ureaplasma parvum. Am J Physiol Lung Cell Mol Physiol. 2010;299:L852–860. doi: 10.1152/ajplung.00183.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gantert M, Jellema RK, Heineman H, Gantert J, Collins JJ, Seehase M, Lambermont VA, Keck A, Garnier Y, Zimmermann LJ, Kadyrov M, Gavilanes AW, Kramer BW. Lipopolysaccharide-induced chorioamnionitis is confined to one amniotic compartment in twin pregnant sheep. Neonatology. 2012;102:81–88. doi: 10.1159/000338015. [DOI] [PubMed] [Google Scholar]
- 93.Debillon T, Gras-Leguen C, Leroy S, Caillon J, Roze JC, Gressens P. Patterns of cerebral inflammatory response in a rabbit model of intrauterine infection-mediated brain lesion. Brain Res Dev Brain Res. 2003;145:39–48. doi: 10.1016/s0165-3806(03)00193-7. [DOI] [PubMed] [Google Scholar]
- 94.Harju K, Ojaniemi M, Rounioja S, Glumoff V, Paananen R, Vuolteenaho R, Hallman M. Expression of toll-like receptor 4 and endotoxin responsiveness in mice during perinatal period. Pediatr Res. 2005;57:644–648. doi: 10.1203/01.PDR.0000156212.03459.A9. [DOI] [PubMed] [Google Scholar]
- 95.Kallapur SG, Kramer BW, Nitsos I, Pillow JJ, Collins JJ, Polglase GR, Newnham JP, Jobe AH. Pulmonary and systemic inflammatory responses to intra-amniotic IL-1alpha in fetal sheep. Am J Physiol Lung Cell Mol Physiol. 2011;301:L285–295. doi: 10.1152/ajplung.00446.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boles JL, Ross MG, Beloosesky R, Desai M, Belkacemi L. Placental-mediated increased cytokine response to lipopolysaccharides: a potential mechanism for enhanced inflammation susceptibility of the preterm fetus. J Inflamm Res. 2012;5:67–75. doi: 10.2147/JIR.S32108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waldorf KM, Gravett MG, McAdams RM, Paolella LJ, Gough GM, Carl DJ, Bansal A, Liggitt HD, Kapur RP, Reitz FB, Rubens CE. Choriodecidual group B streptococcal inoculation induces fetal lung injury without intra-amniotic infection and preterm labor in Macaca nemestrina. PLoS One. 2011;6:e28972. doi: 10.1371/journal.pone.0028972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seehase M, Gantert M, Ladenburger A, Garnier Y, Kunzmann S, Thomas W, Wirbelauer J, Speer CP, Kramer BW. Myocardial response in preterm fetal sheep exposed to systemic endotoxinaemia. Pediatr Res. 2011;70:242–246. doi: 10.1203/PDR.0b013e318225fbcb. [DOI] [PubMed] [Google Scholar]
- 99.Bieghs V, Vlassaks E, Custers A, van Gorp PJ, Gijbels MJ, Bast A, Bekers O, Zimmermann LJ, Lutjohann D, Voncken JW, Gavilanes AW, Kramer BW, Shiri-Sverdlov R. Chorioamnionitis induced hepatic inflammation and disturbed lipid metabolism in fetal sheep. Pediatr Res. 2010;68:466–472. doi: 10.1203/PDR.0b013e3181f70eeb. [DOI] [PubMed] [Google Scholar]
- 100.Grigsby PL, Novy MJ, Adams Waldorf KM, Sadowsky DW, Gravett MG. Choriodecidual inflammation: a harbinger of the preterm labor syndrome. Reprod Sci. 2010;17:85–94. doi: 10.1177/1933719109348025. [DOI] [PubMed] [Google Scholar]
- 101.Kallapur SG, Nitsos I, Moss TJ, Polglase GR, Pillow JJ, Cheah FC, Kramer BW, Newnham JP, Ikegami M, Jobe AH. IL-1 mediates pulmonary and systemic inflammatory responses to chorioamnionitis induced by lipopolysaccharide. Am J Respir Crit Care Med. 2009;179:955–961. doi: 10.1164/rccm.200811-1728OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kannan S, Saadani-Makki F, Muzik O, Chakraborty P, Mangner TJ, Janisse J, Romero R, Chugani DC. Microglial activation in perinatal rabbit brain induced by intrauterine inflammation: detection with 11C-(R)-PK11195 and small-animal PET. J Nucl Med. 2007;48:946–954. doi: 10.2967/jnumed.106.038539. [DOI] [PubMed] [Google Scholar]
- 103.Garnier Y, Coumans AB, Berger R, Hasaart TH. Pulmonary perfusion during lipopolysaccharide (LPS) induced fetal endotoxemia in the preterm fetal sheep. Eur J Obstet Gynecol Reprod Biol. 2006;124:150–157. doi: 10.1016/j.ejogrb.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 104.Rounioja S, Rasanen J, Glumoff V, Ojaniemi M, Makikallio K, Hallman M. Intra-amniotic lipopolysaccharide leads to fetal cardiac dysfunction. A mouse model for fetal inflammatory response. Cardiovasc Res. 2003;60:156–164. doi: 10.1016/s0008-6363(03)00338-9. [DOI] [PubMed] [Google Scholar]
- 105.Burd I, Balakrishnan B, Kannan S. Models of fetal brain injury, intrauterine inflammation, and preterm birth. Am J Reprod Immunol. 2012;67:287–294. doi: 10.1111/j.1600-0897.2012.01110.x. [DOI] [PubMed] [Google Scholar]
- 106.Yang Q, El-Sayed Y, Rosenberg-Hasson Y, Hirschberg DL, Nayak NR, Schilling J, Madan A. Multiple cytokine profile in plasma and amniotic fluid in a mouse model of pre-term labor. Am J Reprod Immunol. 2009;62:339–347. doi: 10.1111/j.1600-0897.2009.00743.x. [DOI] [PubMed] [Google Scholar]
- 107.Koga K, Cardenas I, Aldo P, Abrahams VM, Peng B, Fill S, Romero R, Mor G. Activation of TLR3 in the trophoblast is associated with preterm delivery. Am J Reprod Immunol. 2009;61:196–212. doi: 10.1111/j.1600-0897.2008.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cardenas I, Mor G, Aldo P, Lang SM, Stabach P, Sharp A, Romero R, Mazaki-Tovi S, Gervasi M, Means RE. Placental viral infection sensitizes to endotoxin-induced pre-term labor: a double hit hypothesis. Am J Reprod Immunol. 2011;65:110–117. doi: 10.1111/j.1600-0897.2010.00908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thaxton JE, Romero R, Sharma S. TLR9 activation coupled to IL-10 deficiency induces adverse pregnancy outcomes. J Immunol. 2009;183:1144–1154. doi: 10.4049/jimmunol.0900788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Murphy SP, Hanna NN, Fast LD, Shaw SK, Berg G, Padbury JF, Romero R, Sharma S. Evidence for participation of uterine natural killer cells in the mechanisms responsible for spontaneous preterm labor and delivery. Am J Obstet Gynecol. 2009;200:308, e301–309. doi: 10.1016/j.ajog.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wolfs TG, Kallapur SG, Knox CL, Thuijls G, Nitsos I, Polglase GR, Collins JJ, Kroon E, Spierings J, Shroyer NF, Newnham JP, Jobe AH, Kramer BW. Antenatal ureaplasma infection impairs development of the fetal ovine gut in an IL-1-dependent manner. Mucosal Immunol. 2012 doi: 10.1038/mi.2012.97. [DOI] [PubMed] [Google Scholar]
- 112.Gantert M, Kreczmanski P, Kuypers E, Jellema R, Strackx E, Bastian N, Gavilanes AW, Zimmermann LJ, Garnier Y, Schmitz C, Kramer BW. Effects of in utero endotoxemia on the ovine fetal brain: a model for schizophrenia? Front Biosci (Elite Ed) 2012;4:2845–2853. doi: 10.2741/e588. [DOI] [PubMed] [Google Scholar]
- 113.Zhang L, Saito M, Jobe A, Kallapur SG, Newnham JP, Cox T, Kramer B, Yang H, Kemp MW. Intra-amniotic administration of E coli lipopolysaccharides causes sustained inflammation of the fetal skin in sheep. Reprod Sci. 2012;19:1181–1189. doi: 10.1177/1933719112446079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kemp MW, Saito M, Kallapur SG, Jobe AH, Keelan JA, Li S, Kramer B, Zhang L, Knox C, Yaegashi N, Newnham JP. Inflammation of the fetal ovine skin following in utero exposure to Ureaplasma parvum. Reprod Sci. 2011;18:1128–1137. doi: 10.1177/1933719111408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kramer BW. Chorioamnionitis - new ideas from experimental models. Neonatology. 2011;99:320–325. doi: 10.1159/000326620. [DOI] [PubMed] [Google Scholar]
- 116.Gravett MG, Adams KM, Sadowsky DW, Grosvenor AR, Witkin SS, Axthelm MK, Novy MJ. Immunomodulators plus antibiotics delay preterm delivery after experimental intraamniotic infection in a nonhuman primate model. Am J Obstet Gynecol. 2007;197:518, e511–518. doi: 10.1016/j.ajog.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sadowsky DW, Novy MJ, Witkin SS, Gravett MG. Dexamethasone or interleukin-10 blocks interleukin-1beta-induced uterine contractions in pregnant rhesus monkeys. Am J Obstet Gynecol. 2003;188:252–263. doi: 10.1067/mob.2003.70. [DOI] [PubMed] [Google Scholar]
- 118.Gross G, Imamura T, Vogt SK, Wozniak DF, Nelson DM, Sadovsky Y, Muglia LJ. Inhibition of cyclooxygenase-2 prevents inflammation-mediated preterm labor in the mouse. Am J Physiol Regul Integr Comp Physiol. 2000;278:R1415–1423. doi: 10.1152/ajpregu.2000.278.6.R1415. [DOI] [PubMed] [Google Scholar]
- 119.Gervasi MT, Romero R, Bracalente G, Erez O, Dong Z, Hassan SS, Yeo L, Yoon BH, Chaiworapongsa T. Midtrimester amniotic fluid concentrations of interleukin-6 and interferon-gamma-inducible protein-10: evidence for heterogeneity of intra-amniotic inflammation and associations with spontaneous early (<32 weeks) and late (>32 weeks) preterm delivery. J Perinat Med. 2012;40:329–343. doi: 10.1515/jpm-2012-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yoon BH, Romero R, Moon JB, Shim SS, Kim M, Kim G, Jun JK. Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Obstet Gynecol. 2001;185:1130–1136. doi: 10.1067/mob.2001.117680. [DOI] [PubMed] [Google Scholar]
- 121.Yoon BH, Romero R, Kim CJ, Jun JK, Gomez R, Choi JH, Syn HC. Amniotic fluid interleukin-6: a sensitive test for antenatal diagnosis of acute inflammatory lesions of preterm placenta and prediction of perinatal morbidity. Am J Obstet Gynecol. 1995;172:960–970. doi: 10.1016/0002-9378(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 122.Romero R, Yoon BH, Mazor M, Gomez R, Gonzalez R, Diamond MP, Baumann P, Araneda H, Kenney JS, Cotton DB, et al. A comparative study of the diagnostic performance of amniotic fluid glucose, white blood cell count, interleukin-6, and gram stain in the detection of microbial invasion in patients with preterm premature rupture of membranes. Am J Obstet Gynecol. 1993;169:839–851. doi: 10.1016/0002-9378(93)90014-a. [DOI] [PubMed] [Google Scholar]
- 123.Romero R, Yoon BH, Mazor M, Gomez R, Diamond MP, Kenney JS, Ramirez M, Fidel PL, Sorokin Y, Cotton D, et al. The diagnostic and prognostic value of amniotic fluid white blood cell count, glucose, interleukin-6, and gram stain in patients with preterm labor and intact membranes. Am J Obstet Gynecol. 1993;169:805–816. doi: 10.1016/0002-9378(93)90009-8. [DOI] [PubMed] [Google Scholar]
- 124.Romero R, Yoon BH, Kenney JS, Gomez R, Allison AC, Sehgal PB. Amniotic fluid interleukin-6 determinations are of diagnostic and prognostic value in preterm labor. Am J Reprod Immunol. 1993;30:167–183. doi: 10.1111/j.1600-0897.1993.tb00618.x. [DOI] [PubMed] [Google Scholar]
- 125.Romero R, Sepulveda W, Kenney JS, Archer LE, Allison AC, Sehgal PB. Interleukin 6 determination in the detection of microbial invasion of the amniotic cavity. Ciba Found Symp. 1992;167:205–220. doi: 10.1002/9780470514269.ch13. discussion 220–203. [DOI] [PubMed] [Google Scholar]
- 126.Yoon BH, Kim YA, Romero R, Kim JC, Park KH, Kim MH, Park JS. Association of oligohydramnios in women with preterm premature rupture of membranes with an inflammatory response in fetal, amniotic, and maternal compartments. Am J Obstet Gynecol. 1999;181:784–788. doi: 10.1016/s0002-9378(99)70301-7. [DOI] [PubMed] [Google Scholar]
- 127.Ogge G, Romero R, Lee DC, Gotsch F, Than NG, Lee J, Chaiworapongsa T, Dong Z, Mittal P, Hassan SS, Kim CJ. Chronic chorioamnionitis displays distinct alterations of the amniotic fluid proteome. J Pathol. 2011;223:553–565. doi: 10.1002/path.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Redline RW, Heller D, Keating S, Kingdom J. Placental diagnostic criteria and clinical correlation--a workshop report. Placenta. 2005;26 (Suppl A):S114–117. doi: 10.1016/j.placenta.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 129.Kim JS, Romero R, Kim MR, Kim YM, Friel L, Espinoza J, Kim CJ. Involvement of Hofbauer cells and maternal T cells in villitis of unknown aetiology. Histopathology. 2008;52:457–464. doi: 10.1111/j.1365-2559.2008.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gersell DJ, Phillips NJ, Beckerman K. Chronic chorioamnionitis: a clinicopathologic study of 17 cases. Int J Gynecol Pathol. 1991;10:217–229. [PubMed] [Google Scholar]
- 131.Jacques SM, Qureshi F. Chronic chorioamnionitis: a clinicopathologic and immunohistochemical study. Hum Pathol. 1998;29:1457–1461. doi: 10.1016/s0046-8177(98)90016-8. [DOI] [PubMed] [Google Scholar]
- 132.Khong TY, Bendon RW, Qureshi F, Redline RW, Gould S, Stallmach T, Lipsett J, Staples A. Chronic deciduitis in the placental basal plate: definition and interobserver reliability. Hum Pathol. 2000;31:292–295. doi: 10.1016/s0046-8177(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 133.Bartel G, Wahrmann M, Exner M, Regele H, Schillinger M, Horl WH, Bohmig GA. Determinants of the complement-fixing ability of recipient presensitization against HLA antigens. Transplantation. 2007;83:727–733. doi: 10.1097/01.tp.0000256337.18347.aa. [DOI] [PubMed] [Google Scholar]
- 134.Betkowski AS, Graff R, Chen JJ, Hauptman PJ. Panel-reactive antibody screening practices prior to heart transplantation. J Heart Lung Transplant. 2002;21:644–650. doi: 10.1016/s1053-2498(01)00422-3. [DOI] [PubMed] [Google Scholar]
- 135.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 136.Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 137.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhong S, Zong Y, Slikker W., Jr The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 139.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 140.Baid S, Saidman SL, Tolkoff-Rubin N, Williams WW, Delmonico FL, Cosimi AB, Pascual M. Managing the highly sensitized transplant recipient and B cell tolerance. Curr Opin Immunol. 2001;13:577–581. doi: 10.1016/s0952-7915(00)00262-4. [DOI] [PubMed] [Google Scholar]
- 141.Zeevi A, Girnita A, Duquesnoy R. HLA antibody analysis: sensitivity, specificity, and clinical significance in solid organ transplantation. Immunol Res. 2006;36:255–264. doi: 10.1385/IR:36:1:255. [DOI] [PubMed] [Google Scholar]
- 142.Loetscher M, Gerber B, Loetscher P, Jones SA, Piali L, Clark–Lewis I, Baggiolini M, Moser B. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J Exp Med. 1996;184:963–969. doi: 10.1084/jem.184.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Taub DD, Lloyd AR, Conlon K, Wang JM, Ortaldo JR, Harada A, Matsushima K, Kelvin DJ, Oppenheim JJ. Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med. 1993;177:1809–1814. doi: 10.1084/jem.177.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Taub DD, Sayers TJ, Carter CR, Ortaldo JR. Alpha and beta chemokines induce NK cell migration and enhance NK-mediated cytolysis. J Immunol. 1995;155:3877–3888. [PubMed] [Google Scholar]
- 145.Campbell JD, Gangur V, Simons FE, HayGlass KT. Allergic humans are hyporesponsive to a CXCR3 ligand-mediated Th1 immunity-promoting loop. FASEB J. 2004;18:329–331. doi: 10.1096/fj.02-0908fje. [DOI] [PubMed] [Google Scholar]
- 146.Romagnani P, Crescioli C. CXCL10: a candidate biomarker in transplantation. Clin Chim Acta. 2012;413:1364–1373. doi: 10.1016/j.cca.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 147.Tan J, Zhou G. Chemokine receptors and transplantation. Cell Mol Immunol. 2005;2:343–349. [PubMed] [Google Scholar]
- 148.Wysocki CA, Panoskaltsis-Mortari A, Blazar BR, Serody JS. Leukocyte migration and graft-versus-host disease. Blood. 2005;105:4191–4199. doi: 10.1182/blood-2004-12-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Agostini C, Calabrese F, Rea F, Facco M, Tosoni A, Loy M, Binotto G, Valente M, Trentin L, Semenzato G. Cxcr3 and its ligand CXCL10 are expressed by inflammatory cells infiltrating lung allografts and mediate chemotaxis of T cells at sites of rejection. Am J Pathol. 2001;158:1703–1711. doi: 10.1016/S0002-9440(10)64126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Belperio JA, Keane MP, Burdick MD, Lynch JP, 3rd, Xue YY, Li K, Ross DJ, Strieter RM. Critical role for CXCR3 chemokine biology in the pathogenesis of bronchiolitis obliterans syndrome. J Immunol. 2002;169:1037–1049. doi: 10.4049/jimmunol.169.2.1037. [DOI] [PubMed] [Google Scholar]
- 151.Crescioli C, Buonamano A, Scolletta S, Sottili M, Francalanci M, Giomarelli P, Biagioli B, Lisi G, Pradella F, Serio M, Romagnani P, Maccherini M. Predictive role of pretransplant serum CXCL10 for cardiac acute rejection. Transplantation. 2009;87:249–255. doi: 10.1097/TP.0b013e3181919f5d. [DOI] [PubMed] [Google Scholar]
- 152.Fahmy NM, Yamani MH, Starling RC, Ratliff NB, Young JB, McCarthy PM, Feng J, Novick AC, Fairchild RL. Chemokine and receptor-gene expression during early and late acute rejection episodes in human cardiac allografts. Transplantation. 2003;75:2044–2047. doi: 10.1097/01.TP.0000069601.73079.94. [DOI] [PubMed] [Google Scholar]
- 153.Hoffman SA, Wang L, Shah CV, Ahya VN, Pochettino A, Olthoff K, Shaked A, Wille K, Lama VN, Milstone A, Ware LB, Orens J, Weinacker A, Demissie E, Bellamy S, Kawut SM, Hancock WW, Christie JD. Plasma cytokines and chemokines in primary graft dysfunction post-lung transplantation. Am J Transplant. 2009;9:389–396. doi: 10.1111/j.1600-6143.2008.02497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lazzeri E, Rotondi M, Mazzinghi B, Lasagni L, Buonamano A, Rosati A, Pradella F, Fossombroni V, La Villa G, Gacci M, Bertoni E, Serio M, Salvadori M, Romagnani P. High CXCL10 expression in rejected kidneys and predictive role of pretransplant serum CXCL10 for acute rejection and chronic allograft nephropathy. Transplantation. 2005;79:1215–1220. doi: 10.1097/01.tp.0000160759.85080.2e. [DOI] [PubMed] [Google Scholar]
- 155.Lo DJ, Weaver TA, Kleiner DE, Mannon RB, Jacobson LM, Becker BN, Swanson SJ, Hale DA, Kirk AD. Chemokines and their receptors in human renal allotransplantation. Transplantation. 2011;91:70–77. doi: 10.1097/TP.0b013e3181fe12fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Melter M, Exeni A, Reinders ME, Fang JC, McMahon G, Ganz P, Hancock WW, Briscoe DM. Expression of the chemokine receptor CXCR3 and its ligand IP-10 during human cardiac allograft rejection. Circulation. 2001;104:2558–2564. doi: 10.1161/hc4601.098010. [DOI] [PubMed] [Google Scholar]
- 157.Segerer S, Cui Y, Eitner F, Goodpaster T, Hudkins KL, Mack M, Cartron JP, Colin Y, Schlondorff D, Alpers CE. Expression of chemokines and chemokine receptors during human renal transplant rejection. Am J Kidney Dis. 2001;37:518–531. [PubMed] [Google Scholar]
- 158.Rotondi M, Rosati A, Buonamano A, Lasagni L, Lazzeri E, Pradella F, Fossombroni V, Cirami C, Liotta F, La Villa G, Serio M, Bertoni E, Salvadori M, Romagnani P. High pretransplant serum levels of CXCL10/IP-10 are related to increased risk of renal allograft failure. Am J Transplant. 2004;4:1466–1474. doi: 10.1111/j.1600-6143.2004.00525.x. [DOI] [PubMed] [Google Scholar]
- 159.Tanner SM, Austin JL, Leone G, Rush LJ, Plass C, Heinonen K, Mrozek K, Sill H, Knuutila S, Kolitz JE, Archer KJ, Caligiuri MA, Bloomfield CD, de La Chapelle A. BAALC, the human member of a novel mammalian neuroectoderm gene lineage, is implicated in hematopoiesis and acute leukemia. Proc Natl Acad Sci U S A. 2001;98:13901–13906. doi: 10.1073/pnas.241525498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Qian Z, Fernald AA, Godley LA, Larson RA, Le Beau MM. Expression profiling of CD34+ hematopoietic stem/progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proc Natl Acad Sci U S A. 2002;99:14925–14930. doi: 10.1073/pnas.222491799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gramatges MM, Fani P, Nadeau K, Pereira S, Jeng MR. Neonatal alloimmune thrombocytopenia and neutropenia associated with maternal human leukocyte antigen antibodies. Pediatr Blood Cancer. 2009;53:97–99. doi: 10.1002/pbc.21979. [DOI] [PubMed] [Google Scholar]
- 162.Tomicic M, Starcevic M, Bux J, Zach V, Hundric-Haspl Z, Drazic V, Grahovac B. Severe neonatal neutropenia due to anti-human leucocyte antigen B49 alloimmunization only: a case report. Transfus Med. 2003;13:233–237. doi: 10.1046/j.1365-3148.2003.00446.x. [DOI] [PubMed] [Google Scholar]
- 163.Chan DC, Chen MM, Ooi EM, Watts GF. An ABC of apolipoprotein C-III: a clinically useful new cardiovascular risk factor? Int J Clin Pract. 2008;62:799–809. doi: 10.1111/j.1742-1241.2007.01678.x. [DOI] [PubMed] [Google Scholar]
- 164.Kawakami A, Yoshida M. Apolipoprotein CIII links dyslipidemia with atherosclerosis. J Atheroscler Thromb. 2009;16:6–11. doi: 10.5551/jat.e607. [DOI] [PubMed] [Google Scholar]
- 165.Godfrey KM, Barker DJ. Fetal programming and adult health. Public Health Nutr. 2001;4:611–624. doi: 10.1079/phn2001145. [DOI] [PubMed] [Google Scholar]
- 166.Ravelli GP, Stein ZA, Susser MW. Obesity in young men after famine exposure in utero and early infancy. N Engl J Med. 1976;295:349–353. doi: 10.1056/NEJM197608122950701. [DOI] [PubMed] [Google Scholar]
- 167.Stein AD, Zybert PA, van der Pal-de Bruin K, Lumey LH. Exposure to famine during gestation, size at birth, and blood pressure at age 59 y: evidence from the Dutch Famine. Eur J Epidemiol. 2006;21:759–765. doi: 10.1007/s10654-006-9065-2. [DOI] [PubMed] [Google Scholar]
- 168.Roseboom TJ, van der Meulen JH, Osmond C, Barker DJ, Ravelli AC, Schroeder-Tanka JM, van Montfrans GA, Michels RP, Bleker OP. Coronary heart disease after prenatal exposure to the Dutch famine, 1944–45. Heart. 2000;84:595–598. doi: 10.1136/heart.84.6.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lussana F, Painter RC, Ocke MC, Buller HR, Bossuyt PM, Roseboom TJ. Prenatal exposure to the Dutch famine is associated with a preference for fatty foods and a more atherogenic lipid profile. Am J Clin Nutr. 2008;88:1648–1652. doi: 10.3945/ajcn.2008.26140. [DOI] [PubMed] [Google Scholar]
- 170.Colvin RB, Smith RN. Antibody-mediated organ-allograft rejection. Nat Rev Immunol. 2005;5:807–817. doi: 10.1038/nri1702. [DOI] [PubMed] [Google Scholar]
- 171.Le Moine A, Goldman M, Abramowicz D. Multiple pathways to allograft rejection. Transplantation. 2002;73:1373–1381. doi: 10.1097/00007890-200205150-00001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.