Abstract
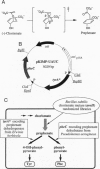
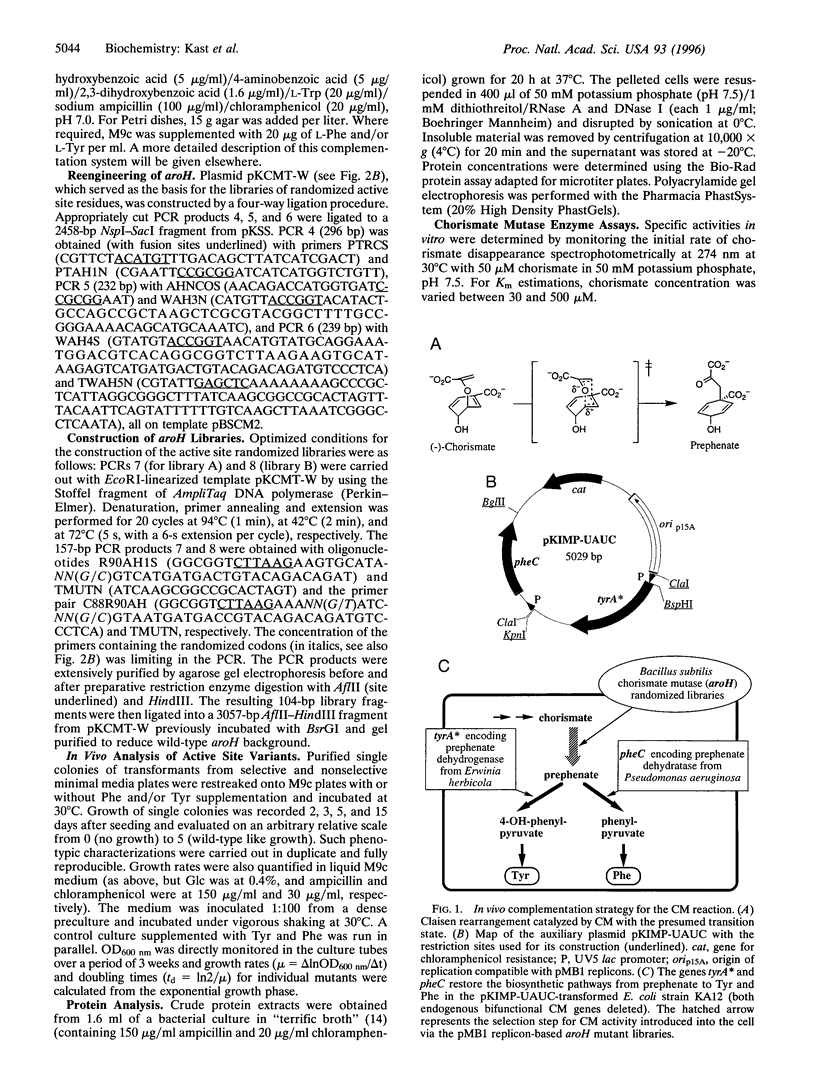
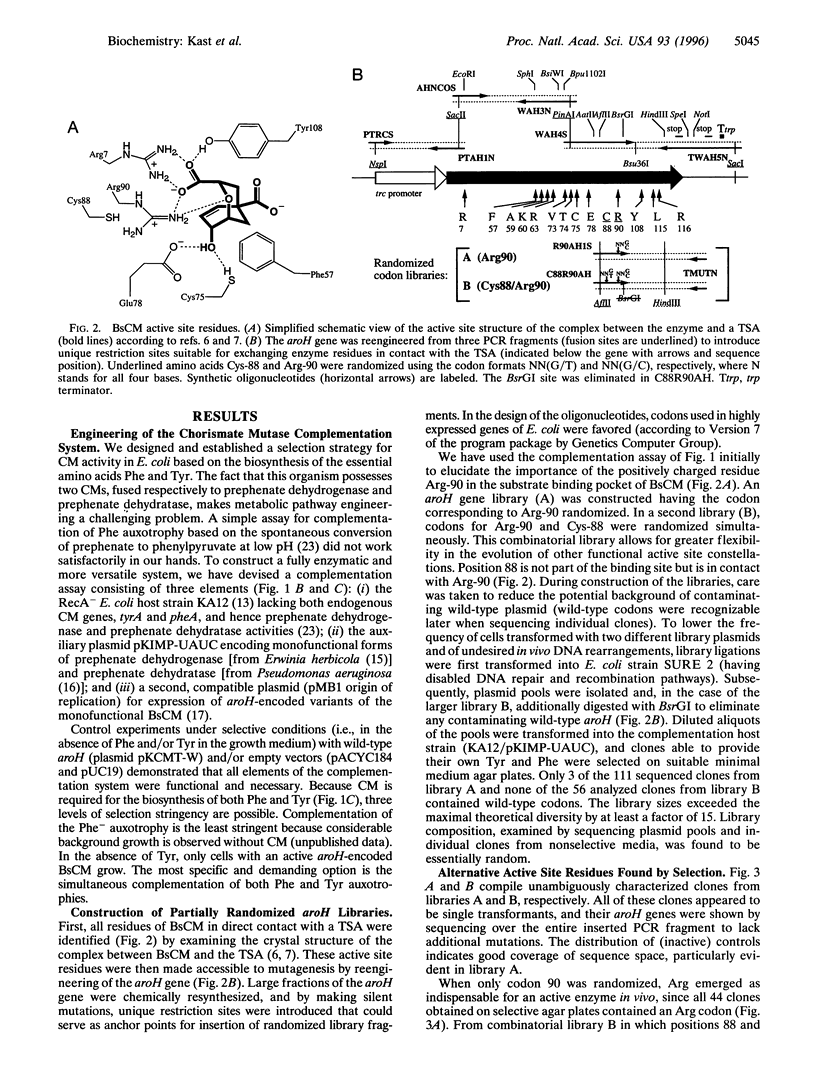
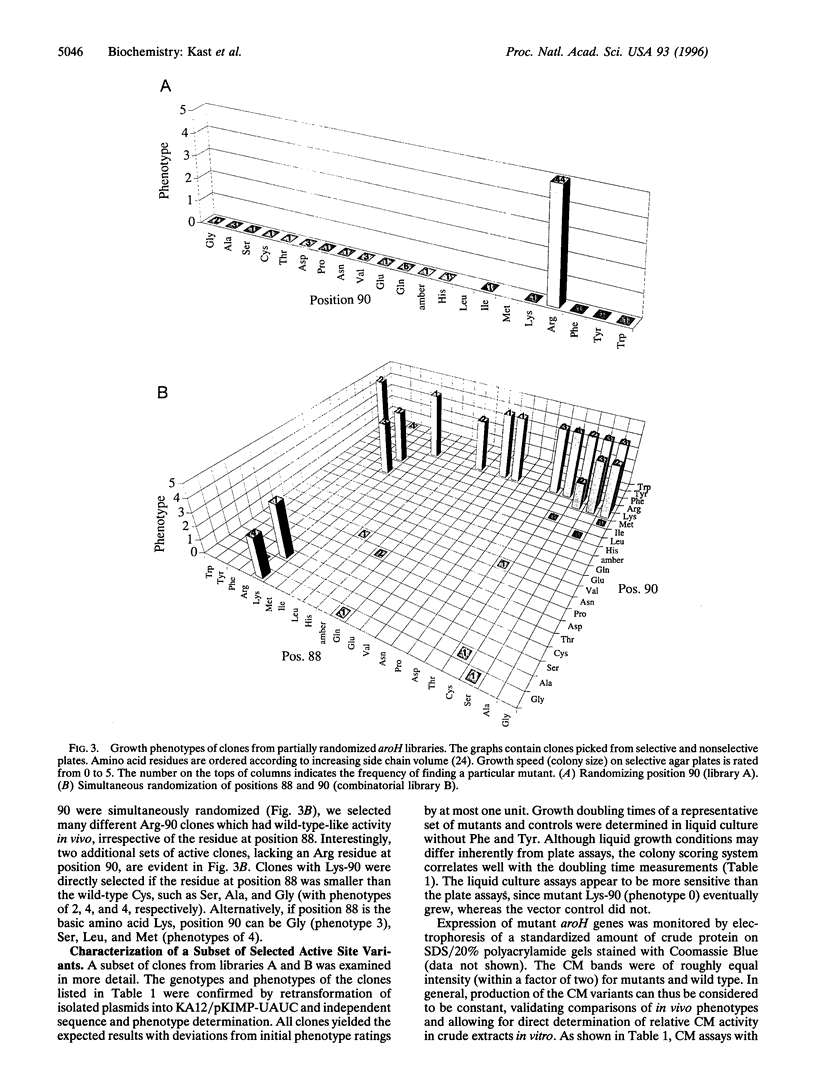
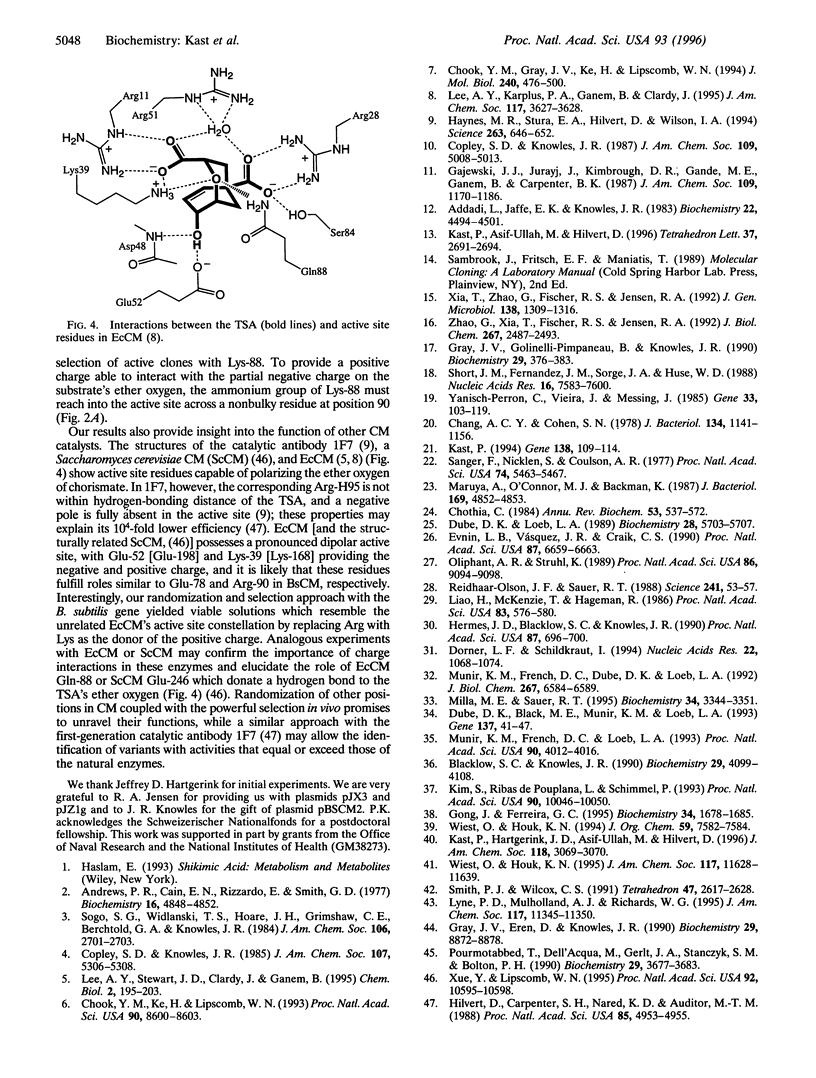
Chorismate mutase (EC 5.4.99.5) catalyzes the intramolecular rearrangement of chorismate to prephenate. Arg-90 in the active site of the enzyme from Bacillus subtilis is in close proximity to the substrate's ether oxygen and may contribute to efficient catalysis by stabilizing the presumed dipolar transition state that would result upon scission of the C--O bond. To test this idea, we have developed a novel complementation system for chorismate mutase activity in Escherichia coli by reengineering parts of the aromatic amino acid biosynthetic pathway. The codon for Arg-90 was randomized, alone and in combination with that for Cys-88, and active clones were selected. The results show that a positively charged residue either at position 88 (Lys) or 90 (Arg or Lys) is essential. Our data provide strong support for the hypothesis that the positive charge is required for stabilization of the transition state of the enzymatic chorismate rearrangement. The new selection system, in conjunction with combinatorial mutagenesis, renders the mechanism of the natural enzyme(s) accessible to further exploration and opens avenues for the improvement of first generation catalytic antibodies with chorismate mutase activity.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Addadi L., Jaffe E. K., Knowles J. R. Secondary tritium isotope effects as probes of the enzymic and nonenzymic conversion of chorismate to prephenate. Biochemistry. 1983 Sep 13;22(19):4494–4501. doi: 10.1021/bi00288a022. [DOI] [PubMed] [Google Scholar]
- Andrews P. R., Cain E. N., Rizzardo E., Smith G. D. Rearrangement of chorismate to prephenate. Use of chorismate mutase inhibitors to define the transition state structure. Biochemistry. 1977 Nov 1;16(22):4848–4852. doi: 10.1021/bi00641a015. [DOI] [PubMed] [Google Scholar]
- Blacklow S. C., Knowles J. R. How can a catalytic lesion be offset? The energetics of two pseudorevertant triosephosphate isomerases. Biochemistry. 1990 May 1;29(17):4099–4108. doi: 10.1021/bi00469a012. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y. M., Gray J. V., Ke H., Lipscomb W. N. The monofunctional chorismate mutase from Bacillus subtilis. Structure determination of chorismate mutase and its complexes with a transition state analog and prephenate, and implications for the mechanism of the enzymatic reaction. J Mol Biol. 1994 Jul 29;240(5):476–500. doi: 10.1006/jmbi.1994.1462. [DOI] [PubMed] [Google Scholar]
- Chook Y. M., Ke H., Lipscomb W. N. Crystal structures of the monofunctional chorismate mutase from Bacillus subtilis and its complex with a transition state analog. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8600–8603. doi: 10.1073/pnas.90.18.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Dorner L. F., Schildkraut I. Direct selection of binding proficient/catalytic deficient variants of BamHI endonuclease. Nucleic Acids Res. 1994 Mar 25;22(6):1068–1074. doi: 10.1093/nar/22.6.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D. K., Black M. E., Munir K. M., Loeb L. A. Selection of new biologically active molecules from random nucleotide sequences. Gene. 1993 Dec 27;137(1):41–47. doi: 10.1016/0378-1119(93)90249-3. [DOI] [PubMed] [Google Scholar]
- Dube D. K., Loeb L. A. Mutants generated by the insertion of random oligonucleotides into the active site of the beta-lactamase gene. Biochemistry. 1989 Jul 11;28(14):5703–5707. doi: 10.1021/bi00440a001. [DOI] [PubMed] [Google Scholar]
- Evnin L. B., Vásquez J. R., Craik C. S. Substrate specificity of trypsin investigated by using a genetic selection. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6659–6663. doi: 10.1073/pnas.87.17.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Ferreira G. C. Aminolevulinate synthase: functionally important residues at a glycine loop, a putative pyridoxal phosphate cofactor-binding site. Biochemistry. 1995 Feb 7;34(5):1678–1685. doi: 10.1021/bi00005a024. [DOI] [PubMed] [Google Scholar]
- Gray J. V., Eren D., Knowles J. R. Monofunctional chorismate mutase from Bacillus subtilis: kinetic and 13C NMR studies on the interactions of the enzyme with its ligands. Biochemistry. 1990 Sep 18;29(37):8872–8878. doi: 10.1021/bi00489a051. [DOI] [PubMed] [Google Scholar]
- Gray J. V., Golinelli-Pimpaneau B., Knowles J. R. Monofunctional chorismate mutase from Bacillus subtilis: purification of the protein, molecular cloning of the gene, and overexpression of the gene product in Escherichia coli. Biochemistry. 1990 Jan 16;29(2):376–383. doi: 10.1021/bi00454a011. [DOI] [PubMed] [Google Scholar]
- Haynes M. R., Stura E. A., Hilvert D., Wilson I. A. Routes to catalysis: structure of a catalytic antibody and comparison with its natural counterpart. Science. 1994 Feb 4;263(5147):646–652. doi: 10.1126/science.8303271. [DOI] [PubMed] [Google Scholar]
- Hermes J. D., Blacklow S. C., Knowles J. R. Searching sequence space by definably random mutagenesis: improving the catalytic potency of an enzyme. Proc Natl Acad Sci U S A. 1990 Jan;87(2):696–700. doi: 10.1073/pnas.87.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilvert D., Carpenter S. H., Nared K. D., Auditor M. T. Catalysis of concerted reactions by antibodies: the Claisen rearrangement. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4953–4955. doi: 10.1073/pnas.85.14.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast P. pKSS--a second-generation general purpose cloning vector for efficient positive selection of recombinant clones. Gene. 1994 Jan 28;138(1-2):109–114. doi: 10.1016/0378-1119(94)90790-0. [DOI] [PubMed] [Google Scholar]
- Kim S., Ribas de Pouplana L., Schimmel P. Diversified sequences of peptide epitope for same-RNA recognition. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10046–10050. doi: 10.1073/pnas.90.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. Y., Stewart J. D., Clardy J., Ganem B. New insight into the catalytic mechanism of chorismate mutases from structural studies. Chem Biol. 1995 Apr;2(4):195–203. doi: 10.1016/1074-5521(95)90269-4. [DOI] [PubMed] [Google Scholar]
- Liao H., McKenzie T., Hageman R. Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc Natl Acad Sci U S A. 1986 Feb;83(3):576–580. doi: 10.1073/pnas.83.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruya A., O'Connor M. J., Backman K. Genetic separability of the chorismate mutase and prephenate dehydrogenase components of the Escherichia coli tyrA gene product. J Bacteriol. 1987 Oct;169(10):4852–4853. doi: 10.1128/jb.169.10.4852-4853.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla M. E., Sauer R. T. Critical side-chain interactions at a subunit interface in the Arc repressor dimer. Biochemistry. 1995 Mar 14;34(10):3344–3351. doi: 10.1021/bi00010a025. [DOI] [PubMed] [Google Scholar]
- Munir K. M., French D. C., Dube D. K., Loeb L. A. Permissible amino acid substitutions within the putative nucleoside binding site of herpes simplex virus type 1 encoded thymidine kinase established by random sequence mutagenesis [corrected]. J Biol Chem. 1992 Apr 5;267(10):6584–6589. [PubMed] [Google Scholar]
- Munir K. M., French D. C., Loeb L. A. Thymidine kinase mutants obtained by random sequence selection. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4012–4016. doi: 10.1073/pnas.90.9.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Struhl K. An efficient method for generating proteins with altered enzymatic properties: application to beta-lactamase. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9094–9098. doi: 10.1073/pnas.86.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourmotabbed T., Dell'Acqua M., Gerlt J. A., Stanczyk S. M., Bolton P. H. Kinetic and conformational effects of lysine substitutions for arginines 35 and 87 in the active site of staphylococcal nuclease. Biochemistry. 1990 Apr 17;29(15):3677–3683. doi: 10.1021/bi00467a013. [DOI] [PubMed] [Google Scholar]
- Reidhaar-Olson J. F., Sauer R. T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science. 1988 Jul 1;241(4861):53–57. doi: 10.1126/science.3388019. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Zhao G., Fischer R. S., Jensen R. A. A monofunctional prephenate dehydrogenase created by cleavage of the 5' 109 bp of the tyrA gene from Erwinia herbicola. J Gen Microbiol. 1992 Jul;138(7):1309–1316. doi: 10.1099/00221287-138-7-1309. [DOI] [PubMed] [Google Scholar]
- Xue Y., Lipscomb W. N. Location of the active site of allosteric chorismate mutase from Saccharomyces cerevisiae, and comments on the catalytic and regulatory mechanisms. Proc Natl Acad Sci U S A. 1995 Nov 7;92(23):10595–10598. doi: 10.1073/pnas.92.23.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhao G. S., Xia T. H., Fischer R. S., Jensen R. A. Cyclohexadienyl dehydratase from Pseudomonas aeruginosa. Molecular cloning of the gene and characterization of the gene product. J Biol Chem. 1992 Feb 5;267(4):2487–2493. [PubMed] [Google Scholar]