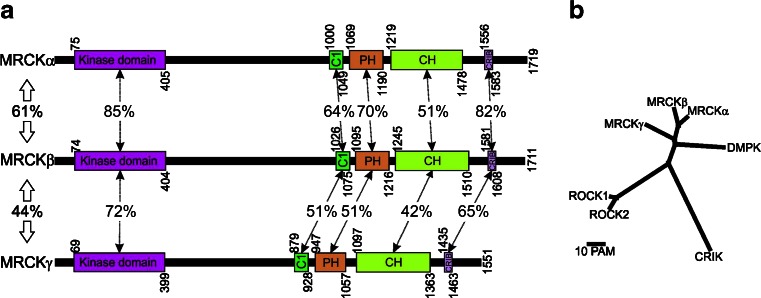
Fig. 1.
Homology between MRCK proteins and related kinases. a Protein domains and their indicated positions were taken from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/protein) for human MRCKα (NP_003598.2), MRCKβ (NP_006026.3) and MRCKγ (NP_059995.2). Percentage amino acid identities were determined with the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi). C1 protein kinase C conserved region 1, PH Pleckstrin homology-like, CH citron homology, CRIB CDC42/Rac interactive binding. b Multiple sequence alignment with hierarchical clustering (http://multalin.toulouse.inra.fr/multalin) was used to create a phylogenetic tree showing the evolutionary relatedness of the kinase domains from MRCK and close homologues. Distance between proteins is depicted by the scale bar, where PAM units are a function of random mutations during evolution that generate diversity