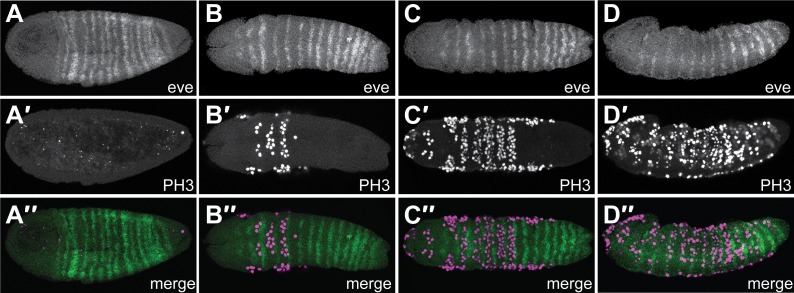
Figure 3. Nv eve expression and cell division appear to be coordinated.
Embryos co-stained for Nv eve mRNA using in situ hybridization and fluorescent detection, as well as for mitotic figures, using an antibody against phospho Histone H3. Embryos are shown with anterior left and dorsal up, except columns B and C, which are ventral views. (A–A″) An early gastrula embryo exhibiting 15 stripes of Nv eve, including five derivatives of stripe 6 (A), has no evident mitotic figures in the posterior domain of Nv eve stripe 6 differentiation (A′). (A″) Merge of panels A and A′. (B–D″) Timecourse series of wild-type embryos stained for Nv eve mRNA and phospho-Histone H3. (B–D). Top panels are Nv eve in situ alone, middle panels (B′–D′) are phospho-Histone H3 antibody staining, and bottom panels (B″–D″) are merge images of upper panels, showing localization of mitotic figures relative to Nv eve stripes.