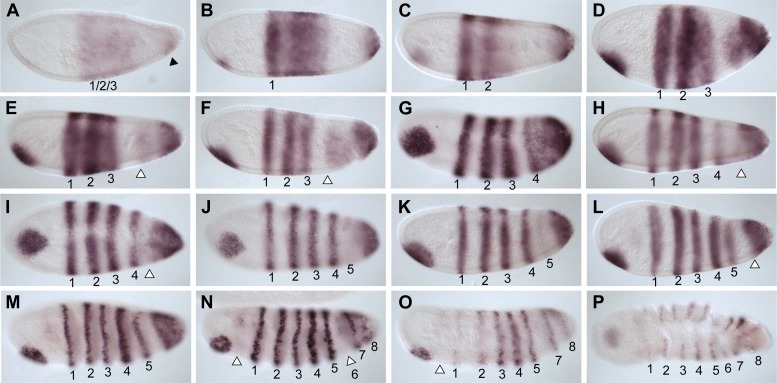
Figure 5. Summary of Nv odd-skipped mRNA expression.
Embryos are shown with anterior left and dorsal up, except where indicated. (A) Precellular blastoderm embryo showing early expression of Nv odd in a broad domain and a posterior cap with a slight clearing in between. (B) Precellular blastoderm embryo showing ventral head patch and darkened central broad domain and distinct posterior cap. (C) Precellular blastoderm embryo with sharpening pair-rule stripes and expanding posterior cap. (D) Precellular blastoderm embryo with dark ventral head patch and posterior cap, and expansion of expression between broad central domain and posterior domain. (E and F) Cellularizing blastoderm embryos with three double-segment periodicity stripes, and a continuous posterior domain of variable staining intensity. Arrowhead indicates boundary of faint expression, which prefigures position of double-segment stripe 4. (G) Ventral view of cellularizing embryo with three strong double-segment stripes, and a fourth stripe forming at the anterior boundary of a more uniformly staining posterior cap (arrowhead). (H) Cellularized blastoderm embryo with four distinct double-segment stripes and a receding posterior cap domain (arrowhead). (I) Ventral view of cellular blastoderm showing four strong double-segment stripes and receding posterior cap (arrowhead), whose anterior boundary prefigures the position of stripe 5. (J) Ventrolateral view of cellular blasoderm embryo showing early appearance of stripe 5 at the anterior boundary of receding posterior domain, whose staining intensity is now less uniform. (K) Cellular blastoderm embryo with five double-segment stripes of expression, a strong ventral head spot, and a reduced, uniform posterior cap. (L) Same as K, with five equivalently strong double-segment stripes. Arrowhead indicates slightly expanded posterior cap. (M) Early germ-band extension embryo with five double-segment periodicity stripes and two stripes becoming evident within the posterior cap. (N) Slightly later embryo than M, with 2 posterior cap stripes more clearly differentiated. (O) Slightly later embryo than N, with anterior stripes fading and posterior segments expanding. (P) Dorsal view, dorsal closure embryo exhibiting eight single-segment periodicity stripes.