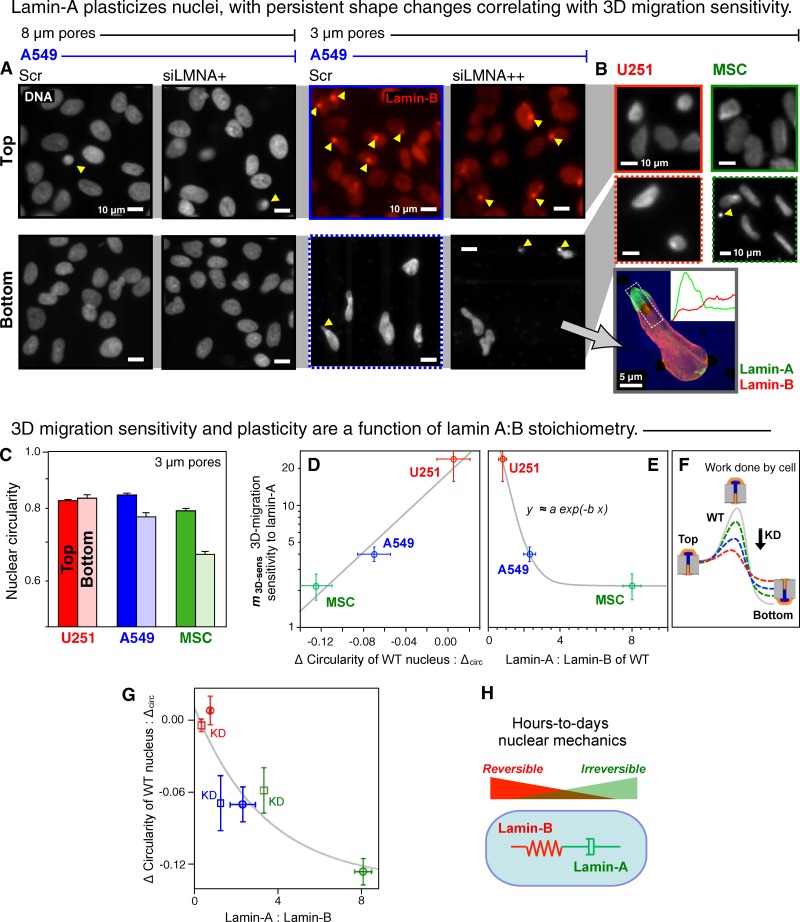
Figure 2.
Lamin-A plasticizes nuclei, with persistent shape changes correlating with both 3D migration sensitivity to lamin-A and also with the lamin-A:B ratio. (A) A549 nuclei on the filters stained for either lamin-B or DNA. Arrowheads indicate a nucleus in a pore. Knockdown with siLMNA was >95% (3 µm) or 60% (8 µm). Inset: confocal image of siLMNA cell on the bottom of 3-µm pore filter showing segregation of lamins, with intensity profiles quantified from z-stacks. (B) U251 and MSC cells imaged on the top and bottom of 3-µm pore filters. (C) Circularity of wild-type nuclei on the top versus bottom of 3-µm filters (log scale). (D) Exponential correlation between 3D migration sensitivity to lamin-A levels (values in Fig. 1 C) and the difference in circularity (n > 3 repeats; ±SEM). The three datapoints were fitted to an exponential, y = α exp(−β x1) with two parameters: α = 19, β = 19 (R2 > 0.95). (E) For 3D migration sensitivity to lamin-A:B ratio, we fit a similar exponential y = a exp(−b x2) + c with similar fit parameters: a (∼α) = 80, b = 2, c << a (R2 > 0.94). (F) Scheme illustrates migration through a pore as an activated process in which an equi-mass knockdown of lamin-A suppresses the barrier and facilitates migration as U251 > A549 > MSC. (G) Migration-induced nuclear circularity change is also nonlinear in lamin-A:B for both wild-type and knockdown cells (KD, squares), with a fit based on the previous fits. (H) Schematic illustrates distinct physical roles of lamin-A in nuclear viscosity (dash pot) and lamin-B in nuclear elasticity (spring).