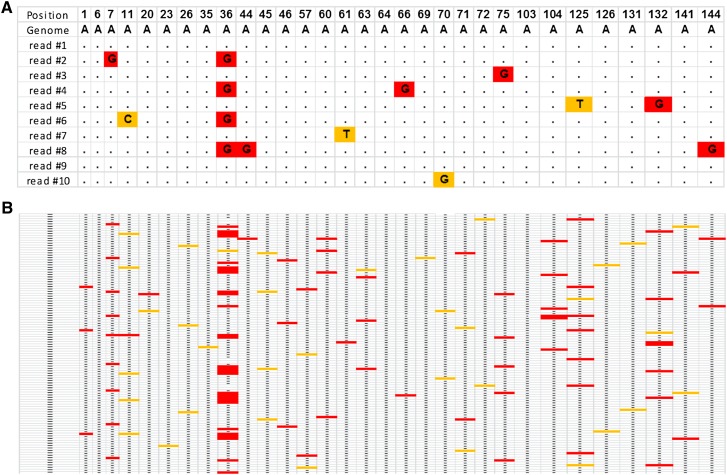
Figure 1.
Detection of A-to-I editing in Alu repeats. (A) Multiple alignment of reads to the reference genome reveals sites of A-to-I editing (red), as well as genomic polymorphisms and sequencing errors (yellow). Detection sensitivity is improved upon examining clusters of mismatches rather than looking at each site independently. Yet, at low coverage, many bona fide editing sites either do not show any AG mismatch, or show a weak signal indistinguishable from sequencing errors. The sites detected include the few strongly edited sites and a random sample of the weaker sites. (B) Ultradeep coverage enables the full scope of editing to be revealed, showing all sites that support editing, typically at very low levels (<1%).