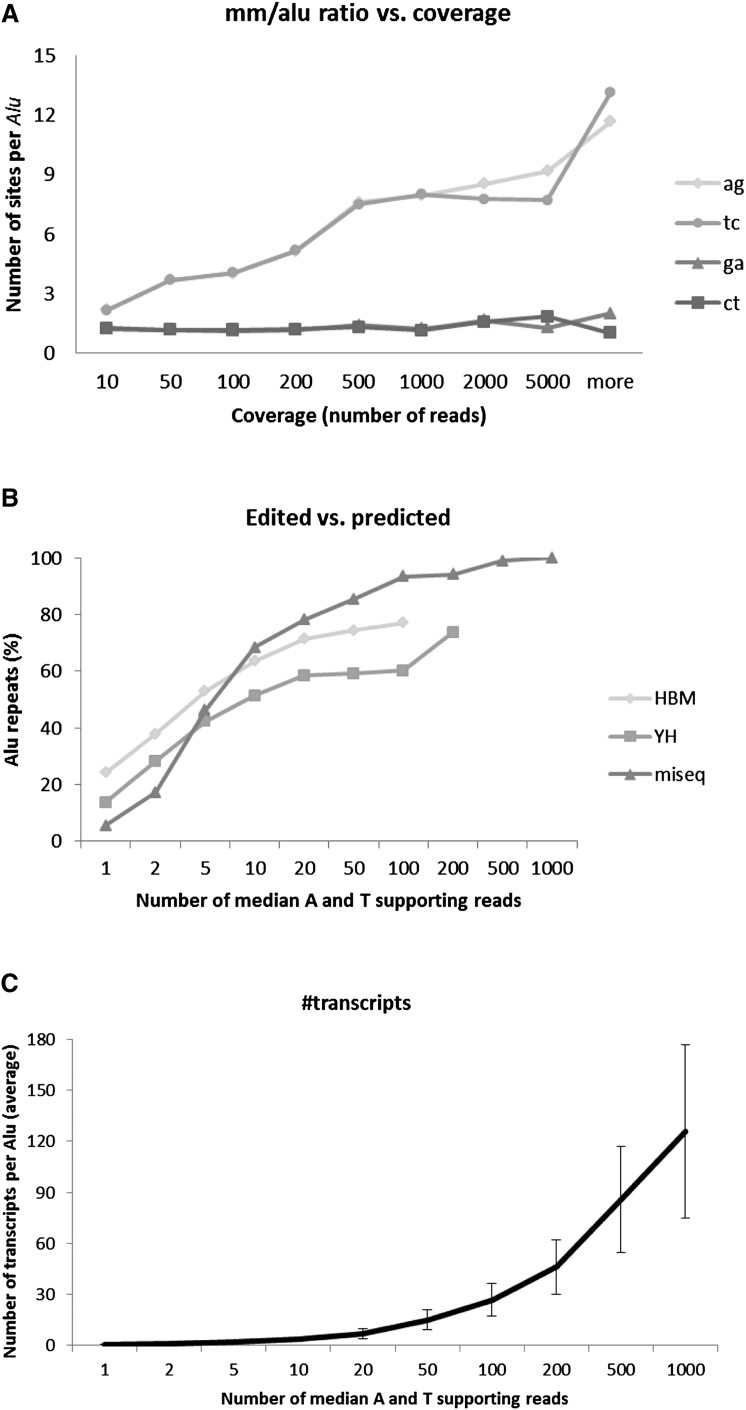
Figure 6.
Editing detection is sensitive to sequencing coverage. (A) The average number of adenosines in an Alu repeat showing evidence for editing increases with the available coverage (number of reads supporting the examined nucleotide), with no sign of saturation (HBM data). A number of mismatch sites of types other than AG/TC saturate at a relatively low coverage (after applying the statistical model to filter sequencing errors). As the typical coverage in RNA-seq is much lower than 1000 reads, this suggests that previous counts of editing sites are grossly underestimated. (B) Fraction of Alu repeats showing evidence of editing (i.e., dominated by AG/TC mismatches). Again, strong dependence on coverage is observed, and atypically high coverage is required for detection in most of the Alu repeats. Our ultradeep MiSeq experiment reached saturation with all Alu repeats detected at a coverage of 1000 reads (coverage is defined as the median read coverage for the adenosines and thymines in the given Alu repeat). Based on these calculations, we estimate the total number of A-to-I editing sites in the human genome to exceed 100 million sites. (C) Number of different transcript variants per Alu, as a function of the reads' coverage. No saturation is observed even for ultrahigh coverage.