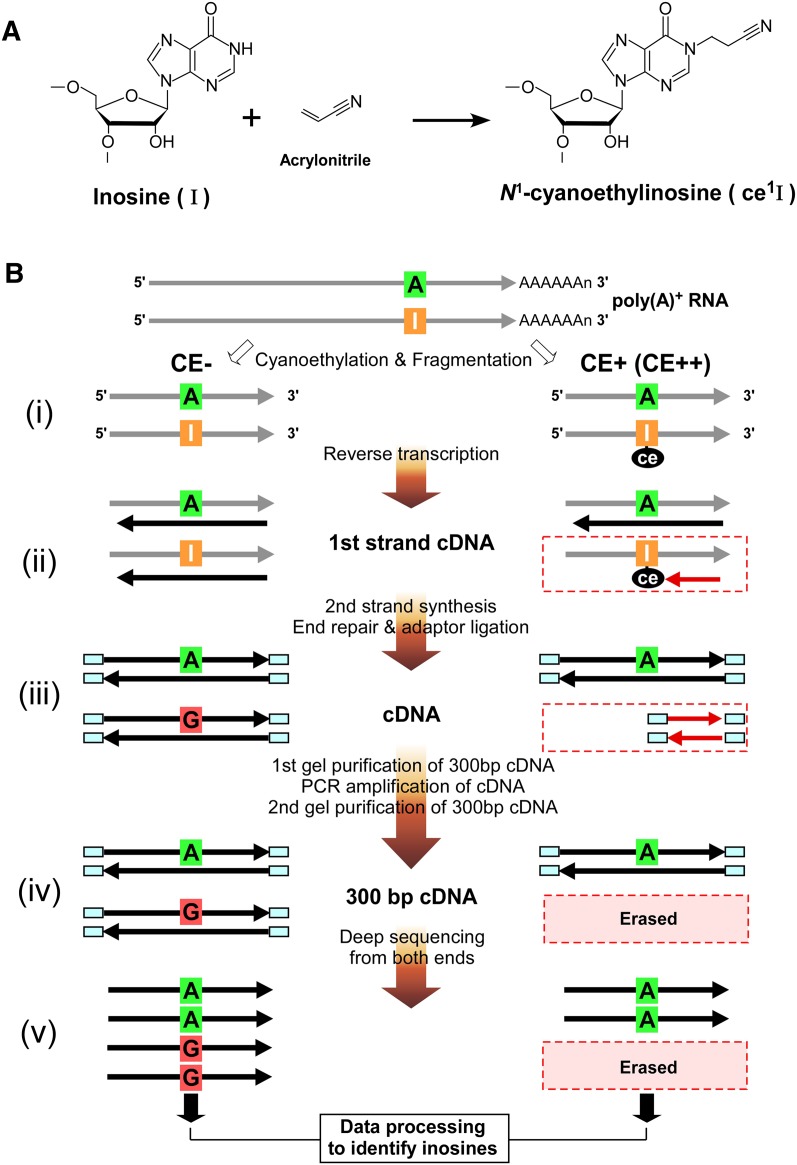
Figure 1.
Biochemical identification of A-to-I editing sites by ICE-seq. (A) Chemistry of inosine cyanoethylation. Acrylonitrile adducts to the N1 position of inosine to form N1-cyanoethylinosine (ce1I). (B) Outline of ICE-seq. Schemes without (CE−) or with (CE+ or CE++) cyanoethylation of RNA are shown on the left and right, respectively. RNA and cDNA are indicated by gray and black arrows, respectively. (i) Cyanoethylation and fragmentation. The I in the RNA strand is specifically cyanoethylated to form ce1I (CE+). RNAs are partially digested by mild alkaline treatment. (ii) First strand cDNA synthesis. RNAs are reverse-transcribed with a random primer. RNA bearing an A at the editing site is converted to T in the cDNA in both conditions (CE− or CE+). In the CE− condition, RNA bearing I is transcribed to C in the cDNA. In the CE+ condition, first strand cDNA extension is arrested at the ce1I site (red arrow). (iii) Second strands are synthesized to obtain double-stranded cDNAs which are then subjected to the end-repair reaction and adaptor ligation. (iv) Gel purification of 300-bp cDNAs for PCR amplification. The amplified cDNAs with 300 bp are gel-purified again. In this step, cDNAs arrested at ce1I are discarded. (v) The cDNAs for CE−, CE+, and CE++ conditions are sequenced from both ends by a GA2 sequencer. Data processing of these reads identifies inosines by detecting erased reads upon cyanoethylation.