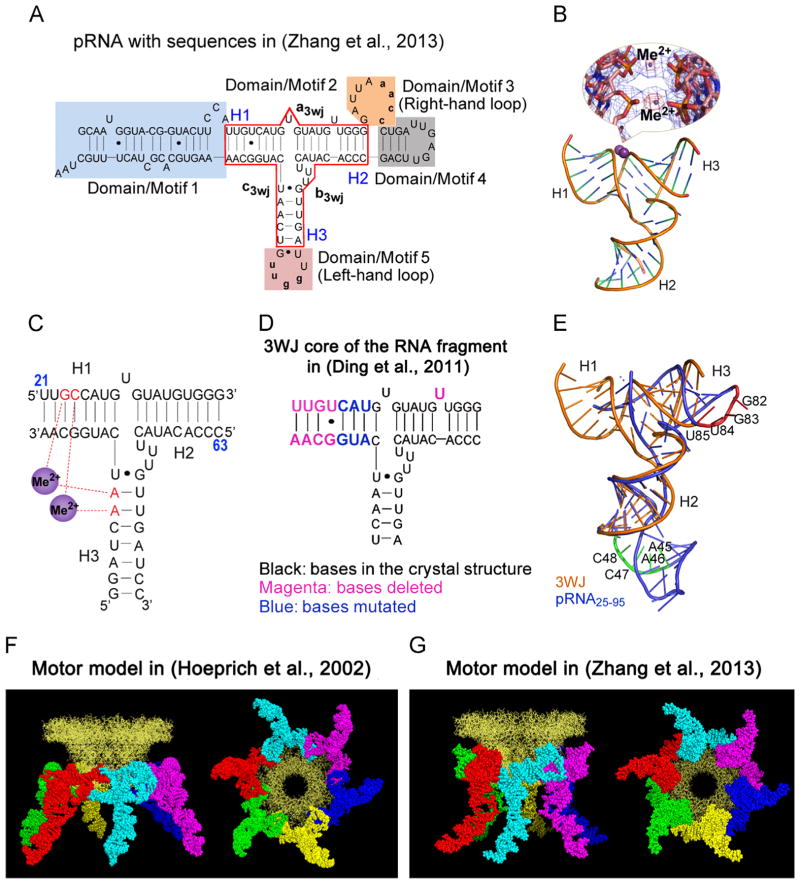
Fig. 2.
Comparison of the two RNA molecules used in crystallization to produce hexamer and pentamer. (A) The sequence of the hexameric pRNA used in (Zhang et al., in press). (B) Crystal structure of pRNA 3WJ with two metal binding sites (Me2+, magenta), shown as sphere. A close-up of the metal binding sites superposed on the 2Fo-Fc electron density map (blue mesh contoured at 1.0 σ) and the anomalous difference map (red mesh contoured at 4.5 σ) are shown in the inset. (C) Schematic representation of the 3WJ structure with metal coordinating nucleotides in red. Numbers in blue represent the nucleotide locations in the wild-type pRNA sequence. (D) The 3WJ core in the RNA fragment sequence used in (Ding et al., 2011), the mutated and deleted bases are shown in different colors as indicated. (E) Superposition of the pRNA25–95 (Ding et al., 2011) (blue) and 3WJ domain (Zhang et al., in press) (gold) crystal structures with the left- (red) and right-hand (green) loops highlighted. The motor model with the pRNA derived from computational and biochemical methods in (Hoeprich and Guo, 2002) (F) and the one derived from crystallization in (Zhang et al., in press) (G) are shown as side view (left) and bottom view (right). (A)–(E) and (G), adapted from Zhang et al. (in press), © 2013 with permission from Cold Spring Harbor Laboratory Press; (F), adapted from Hoeprich and Guo, (2002), © 2002 with permission from The American Society for Biochemistry and Molecular Biology. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)