Figure 1.
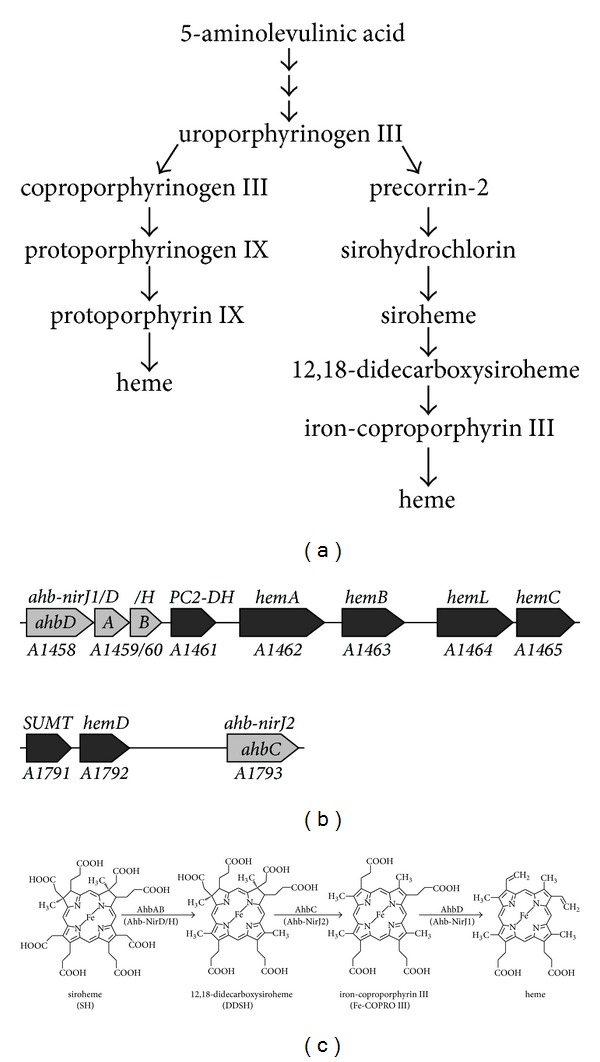
The alternative heme biosynthesis pathway in Methanosarcina barkeri. (a) Comparison of the classical heme biosynthesis pathway (left hand side) with the alternative pathway (right hand side). In both pathways heme is derived from the common precursor uroporphyrinogen III. In the alternative pathway heme is generated via precorrin-2 requiring a total of six consecutive enzymatic steps. In the classical pathway uroporphyrinogen III is converted into heme via coproporphyrinogen III in four reaction steps. (b) Heme biosynthesis gene clusters in M. barkeri. Genes encoding enzymes which catalyze the early steps during heme formation are shown in black. Genes which were proposed to be important for the last three steps of the alternative heme biosynthesis pathway are shown in grey [3]. The gene numbers for M. barkeri Fusaro (Mbar_) are given below the arrows, the original gene designations [3] are given above the arrows, and the newly suggested names for the genes required for the last three steps are indicated in the grey arrows [6]. (c) The last three steps of the alternative heme biosynthesis pathway. First, the two acetate side chains at positions C-12 and C-18 of siroheme are decarboxylated to methyl groups by AhbAB resulting in 12,18-didecarboxysiroheme. AhbC removes the acetate side chains at positions C-2 and C-7 to form iron-coproporphyrin III which is finally converted into heme by AhbD through the oxidative decarboxylation of the propionate side chains at positions C-3 and C-8 to the corresponding vinyl groups. The newly suggested enzyme names are given above the arrows [6], and the original enzyme designations are given below the arrows in parentheses [3].
