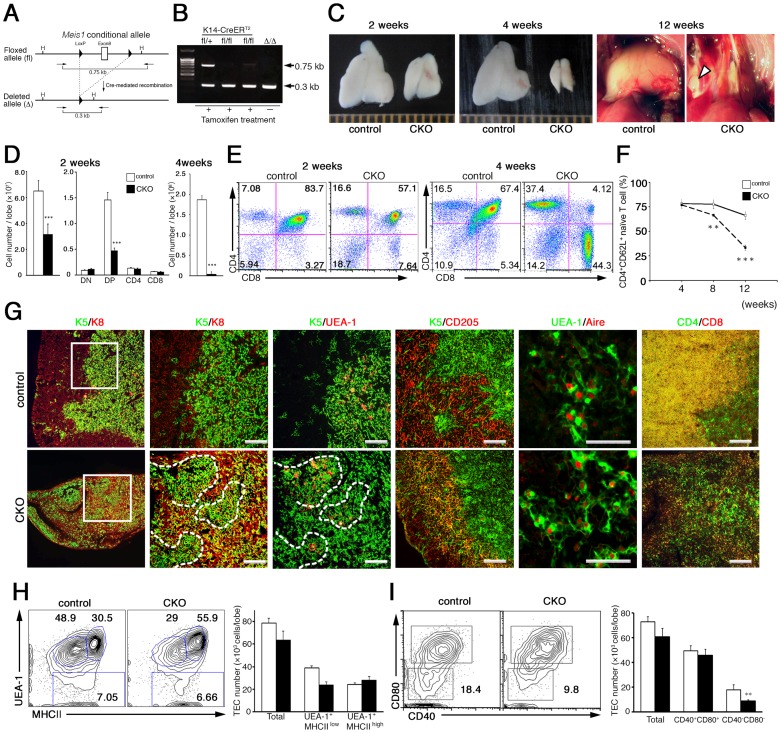
Figure 2. Epithelial-specific deletion of Meis1 causes thymic atrophy.
(A) A diagram depicting the floxed Meis1 allele. LoxP sites (arrowheads) were inserted into intronic sites flanking exon 8 of the Meis1 gene. PCR primers for verifying the Cre-mediated deletion of the loxP-flanked fragment are indicated by arrows. (B) Confirmation of Meis1 deletion in K14-CreERT2 mice. DNAs from sorted EpCAM+ CD205− TECs from K14-CreERT2 Meis1 fl/fl and Meis1 fl/+ mice that were either treated (+) or untreated (−) with tamoxifen were subjected to PCR analysis using primer pairs shown in A. DNAs from Meis1 Δ/Δ mice were used as controls. (C) Gross appearance of thymus lobes from CKO and control mice 2 weeks (left) and 4 weeks (middle) post induction of Meis1 deletion. A macroscopic view of a mediastinum of CKO and control mice 12 weeks post induction of Meis1 deletion is also shown (right). The thymus lobes of control mice were positioned above the heart, whereas in the CKO the thymic lobes had completely disappeared. White arrowheads indicate ectopic lymph nodes. (D) Total cell numbers of total thymocytes (left) and subpopulations of thymocytes at different developmental stages (middle) from CKO (black bars) and control (white bars) mice 2 weeks post induction of Meis1 deletion. Numbers of total thymocytes (right) from CKO (black bars) and control (white bars) mice 4 weeks post induction of Meis1 deletion are also shown. Data are the mean and standard deviation of three independent experiments. ***, p<0.001. (E) Representative FACS profiles of thymocyte differentiation in CKO and control mice 2 weeks (left two panels) and 4 weeks (right two panels) post induction of Meis1 deletion. (F) The proportion of CD44− CD62L+ naïve CD4 T cells among total CD4 T cells in the peripheral blood of CKO (dotted line) and control (solid line) mice. Data are the mean and standard deviations of three independent experiments. **, p<0.01 and ***, p<0.001. (G) Immunohistochemical analysis of the thymus from CKO and control mice 4 weeks post induction of Meis1 deletion. Tissue sections were double stained with the indicated antibody combinations. Broken lines indicate cortico-medullary borders. Data are representative of 3 independent experiments. Scale bars, 100 µm. (H–I) Representative FACS profiles of TECs from K14-CreERT2 and control mice 4 days after tamoxifen treatment. Numbers indicate percentage of cells within the indicate gates. Bars show the cell number of the indicated EpCAM+ TECs from CKO (black) and control (white) mice. Shown are the mean and standard deviation from three independent experiments. **, p<0.01.