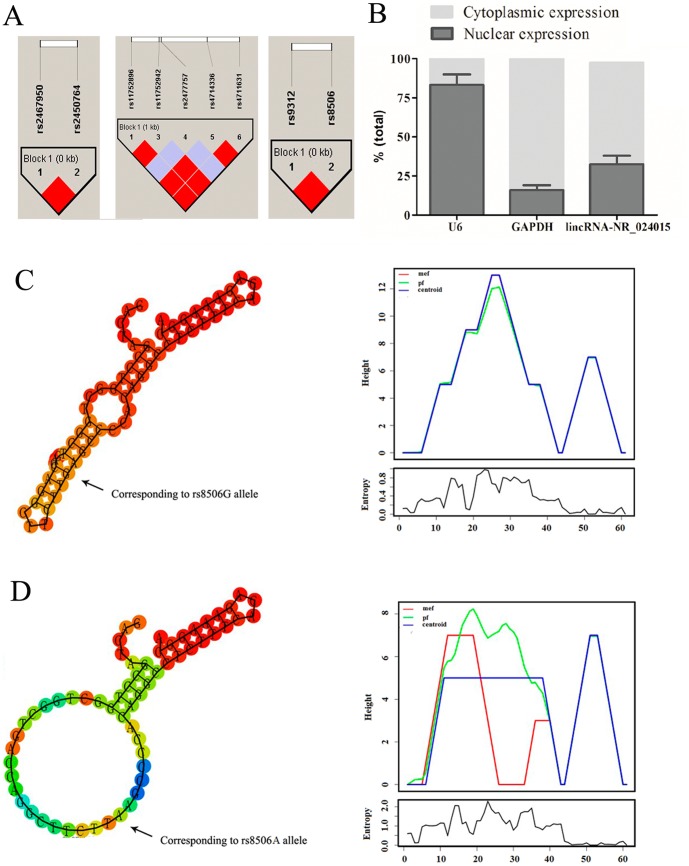
Figure 1. Cellular and molecular characterization of lincRNA-NR_024015.
(A) Linkage disequilibrium (LD) map for selected SNPs of lincRNA-NR_024015 exon. It showed that rs11752896, rs11752942, rs4714336 and rs4711631 are in LD with D′ = 1; rs2467950 and rs2450764 are in LD with D′ = 1; rs9312 and rs8506 are in LD with D′ = 1. (B) The levels of nuclear control transcript (U6), cytoplasmic control transcript (GAPDH mRNA), and lincRNA-NR_024015 were assessed by RT-qPCR in nuclear and cytoplasmic fractions. Data are mean±standard error of the mean. Data are presented as a percentage of U6, GAPDH and lincRNA-NR_024015 levels and total levels for each were taken to be 100%. (C–D) In-silico prediction of folding structures induced by rs8506G>A in lincRNA-NR_024015. The mountain plot is an xy-graph that represents a secondary structure including MFE structure, the thermodynamic ensemble of RNA structures (pf), and the centroid structure in a plot of height versus position. “mfe” represents minimum free energy structure; “pf” indicates partition function; “centroid” represents the centroid structure.