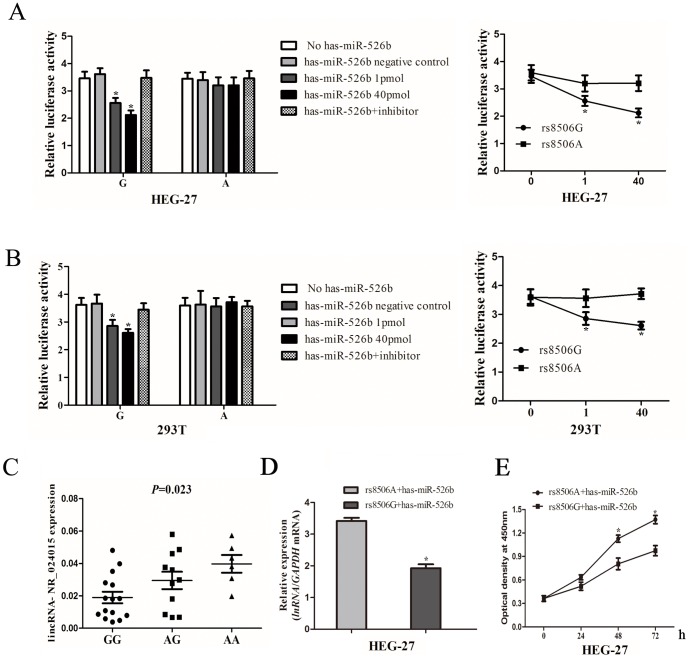
Figure 2. The rs8506G>A genotypes affect lincRNA-NR_024015 expression.
Representative graph of luciferase activity of variant allele on luciferase reporter genes bearing the lincRNA-NR_024015 exonic region spanning 160 bp flanking the rs8506G>A polymorphism segments in HEG-27 (A) and 293T cells (B). Results are shown as percentage relative to luciferase activity (Renilla luciferase activity was measured and normalized to firefly luciferase). Relative luciferase activity of the psiCHECK-2-lincRNA-NR_024015-G-allele and psiCHECK-2- lincRNA-NR_024015-A-allele constructs cotransfected with has-miR-526b mimic and inhibitor. Six replicates for each group and the experiment repeated at least three times. Data are mean ± SEM. Asterisk indicates a significant change (P<0.001). (C) LincRNA-NR_024015 expression levels in thirty-two non-cardia gastric cancer patients for different rs8506G>A genotypes (15 rs8506GG, 11 rs8506AG and 6 rs8506AA); data are mean±standard error of the mean. (D) HEG-37 cells were cotransfected pcDNA-lincRNA-rs8506G or pcDNA-lincRNA-rs8506A with has-miR-526b. After 24 h, cells were collected, RNA extracted and real time PCR performed. Data are mean±standard error of the mean. “asterisk” represents P<0.05. (E) Cells' proliferation rate was significantly inhibited when cells cotranfected lincRNA-NR_024015 haboring rs8506G allele and has-miR-526b. Cell proliferation was performed by the cell viability assay and the effect became obvious from day 2. Six replicates for each group and the experiment repeated at least three times. Data are mean±standard error of the mean. “asterisk” represents P<0.05.