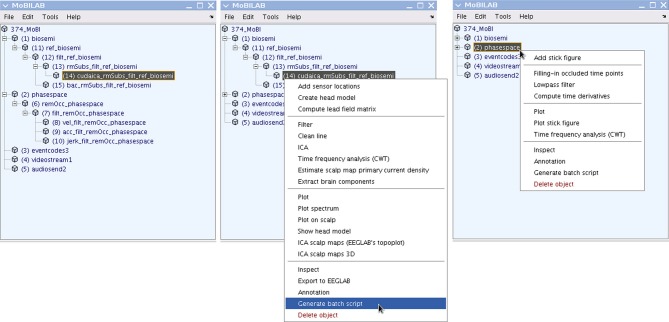
Figure 4.
The MoBILAB GUI and processing pipelines. The left panel shows the tree of parent-children relationships among data objects in a loaded multi-stream MoBI dataset. The integers enclosed in parentheses to the left of each object name give the index in the cell array mobilab.allStream.item. The two branches shown unfolded (by clicking on the data object names, here biosemi and phasespace) represent two already selected data processing pipelines (cf. Text Boxes 1, 2): (1) for Biosemi (Amsterdam) system EEG data: raw data ⇒ re-referencing ⇒ high-pass filtering ⇒ artifact rejection ⇒ ICA decomposition; (2) for PhaseSpace (Inc.) system motion capture data: raw data ⇒ correction of occlusion artifacts ⇒ low-pass filtering ⇒ computation of time derivatives. By following any branch backwards (upwards), the user can generate MATLAB scripts that make it easy to run the same series of operations on other Biosemi and PhaseSpace datasets. The center panel shows the contextually defined menu for the EEG dataStream object. The menu item shaded in blue backtracks the history of every object in the selected branch, creating a script ready to run (see Text Box 1). The right panel shows the contextually defined menu for the motion capture data object (see Text Box 2). Note that the two stream objects (EEG and motion capture) have different processing menus that present their individually defined processing methods.