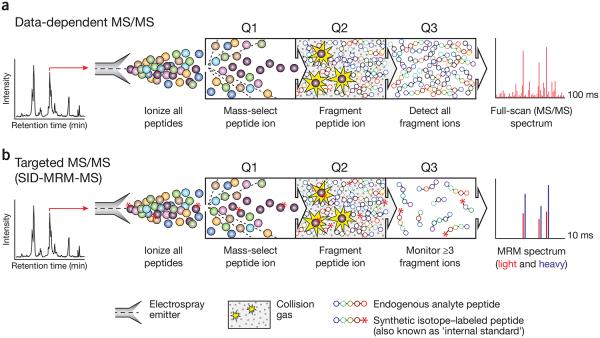
Figure 1.
Comparison of conventional data-dependent analysis to targeted MRM-MS on a triple quadrupole mass spectrometer. (a) In a data-dependent MS experiment, digested proteins are loaded on a reversed-phase column attached to a liquid chromatography setup and eluted via electrospray to yield gas-phase ions. At any given point in the chromatographic separation many tens to hundreds of peptides are eluting nearly simultaneously. A full-scan MS spectrum is acquired and informs collection of subsequent MS/MS scans in which 4–10 ions observed in the MS spectrum are automatically selected on the basis of their signal intensity (Q1) for fragmentation by collision with inert gas (Q2). The complete array of fragment ions is detected (Q3), which constitutes the full-scan MS/MS spectrum (far right). (b) In an SID-MRM-MS analysis, proteotypic peptides uniquely representing proteins of interest are predefined together with their most informative fragment ions. Peptides are selected for fragmentation (Q1 and Q2), and fragment ions are selected for detection (Q3) based on a user-specified list of targeted precursor-fragment pairs (‘transitions’). Synthetic peptides containing stable-isotope labels can be spiked in as standards (asterisks). Comparing labeled to unlabeled peak area (far right) provides precise relative quantification of the endogenous analyte.