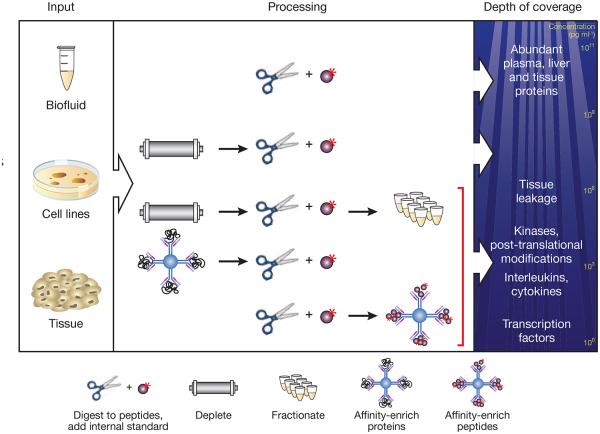
Figure 2.
Protein and peptide enrichment strategies to increase sensitivity and specificity of analyte detection in SID-MRM-MS. After extraction from tissues, cell lines or biofluids (left), proteins are digested into peptides. To achieve the highest detection confidence and measurement precision for peptides in these complex samples, synthetic versions of each analyte peptide containing an amino acid labeled with a stable isotope (for example, 15N or 13C) are added as internal standards (top line, center). Additional strategies may be required to achieve desired detection sensitivity in complex matrices. In plasma, depletion of the 12–70 most abundant plasma proteins using immunoaffinity depletion columns can result in limits of detection of ~100 ng ml−1 (second line, center), representing ‘middling’ depths in the plasma peptide ocean (right). Biomarkers in diseases such as cancer and cardiovascular disease are commonly present in the low nanogram per milliliter range and can be robustly detected by coupling depletion to limited fractionation (third line, center), albeit at the expense of reducing throughput and introducing variability and analytical complexity. A different strategy to enhance sensitivity relies on immunoaffinity reagents to enrich target analytes. In one implementation the intact protein is captured using an anti-protein antibody before digestion (fourth line, center). Alternatively, a proteotypic peptide derived from the protein can be captured using an anti-peptide antibody to the target peptide of interest (bottom line, center).