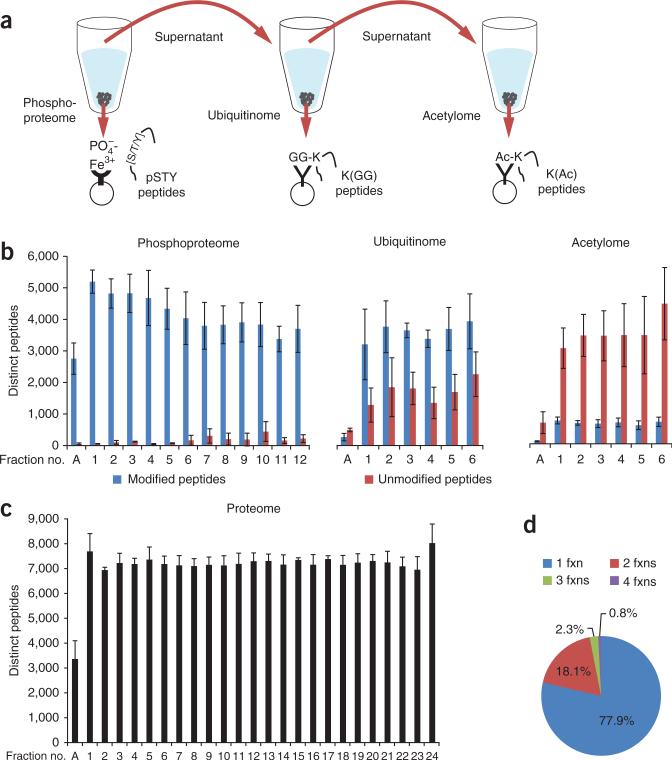
Figure 1.
SEPTM enables deep, quantitative analysis of the PTM-ome of a biological sample. (a) Integrated proteome and PTM-ome profiling by sequential enrichment for phosphorylated peptides with IMAC, for ubiquitinated peptides with K(GG)-specific antibodies and for lysine-acetylated peptides with K(Ac)-specific antibodies. (b) Ideal and even partitioning of phosphorylated, ubiquitinated and acetylated peptides by basic RP fractionation. The specificity of each enrichment step (distinct modified peptides out of total peptides) was >93% for phosphopeptides, >71% for ubiquitinated peptides and >18% for acetylated peptides. (c) We used 5% of the starting samples for whole-proteome analysis; basic RP separation yielded even partitioning of unmodified peptides. Average numbers of PTM sites (b) and proteins (c) per replicate are shown with s.d. error bars. (d) Uniqueness and occurrence of unmodified peptides per fraction (fxn).