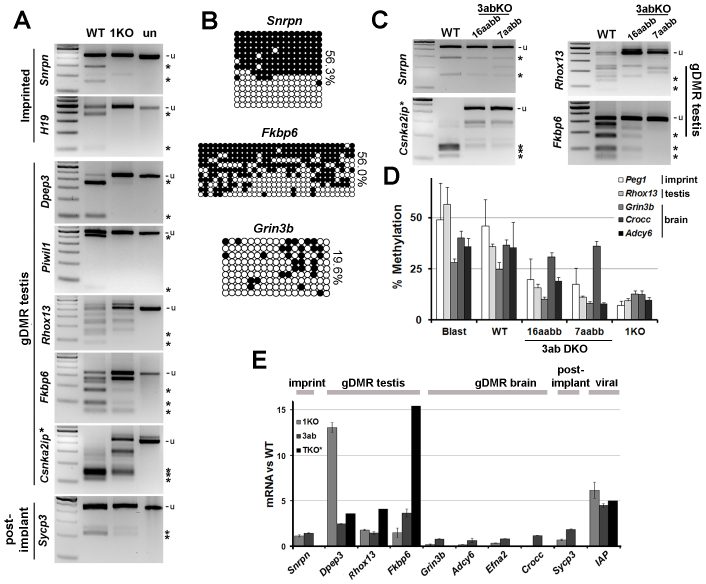
Fig. 5.
Enzyme dependence for gDMR classes in mouse stem cells. (A) Methylation levels in parental J1 ESCs (WT) and DNMT1-deficient derivatives (1KO) assessed by COBRA. un, uncut control. (B) Clonal analysis of a gDMR from an imprinted, testis or brain gene (Snrpn, Fkbp6 and Grin3b, respectively). (C) COBRA showing methylation levels in clonally derived ESCs (16aabb, 7aabb) carrying double knockouts (DKO) in both Dnmt3a and Dnmt3b. (D) Pyrosequencing for the indicated gDMRs. Blastocyst methylation levels are derived from the analysis shown in Fig. 1D. Differences between DKO and WT were significant (P<0.05) for all analysed genes except Crocc. (E) Fold reactivation in mutant versus WT cells as assessed by RT-qPCR. 3ab, DKO cells (as above); TKO, triple KO cells lacking DNMT1, DNMT3A and DNMT3B, data from Karimi et al. (Karimi et al., 2011b); brain and imprint gDMR unavailable. Error bars indicate s.e.m.