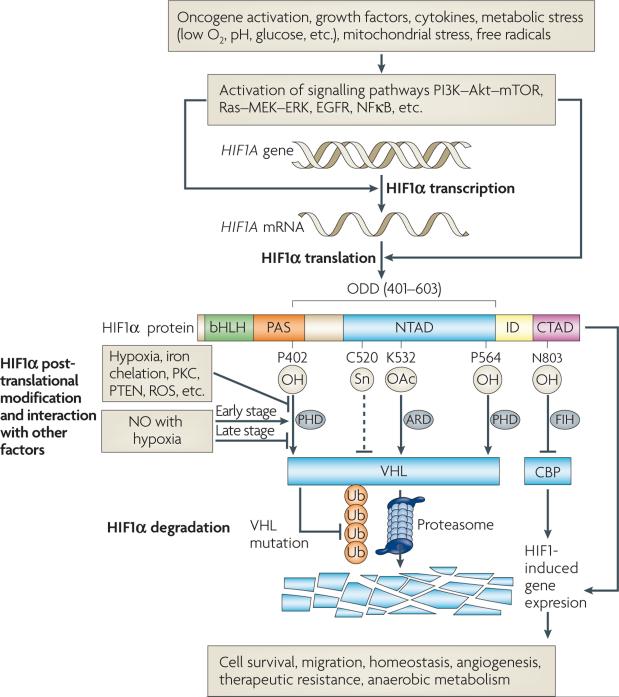
Figure 1. Features of hypoxia-inducible factor (HIF1) regulation.
HIF1 transcriptional activity regulates numerous genes that control a variety of cellular functions, including anaerobic metabolism, angiogenesis stimulation and mechanisms for resistance to therapy. HIF1 transcriptional activity requires formation of a heterodimer consisting of HIF1α and HIF1β. The heterodimer binds to hypoxia response elements (HREs) in promoter regions of its target genes, where it activates transcription. Whereas HIF1β is constitutively expressed, HIF1α protein levels are subject to a number of points of regulation. HIF1α consists of the following regulatory domains: bHLH (basic-helix-loop-helix), PAS (Per-ARNT-Sim), NTAD (N-terminal transactivation domain), CTAD (C-terminal transactivation domain) and ODD (oxygen-dependent degradation domain). The rate of synthesis of HIF1α is controlled by activation of the phosphatidylinositol 3-kinase (PI3K)-Akt and Ras pathways by a variety of stimuli. HIF1α protein is rapidly targeted by the von Hippel-Lindau protein (VHL) complex for proteasomal degradation under normoxic conditions, following an oxygen-dependent prolyl hydroxylation of proline residues in the ODD. Activity of the prolyl hydroxylases (PHDs) is influenced by protein kinase C (PKC), PTEN and reactive oxygen species (ROS). Nitric oxide (NO) has variable effects on the stability of HIF1α, depending on the oxygenation state of the cell. Once HIF1α forms a heterodimer with its partner, HIF1β, the transcriptional activity is further regulated by cofactors, such as CREB binding protein (CBP) or factor-inhibiting HIF (FIH). ARD, acetyl transferase.