Figure 5.
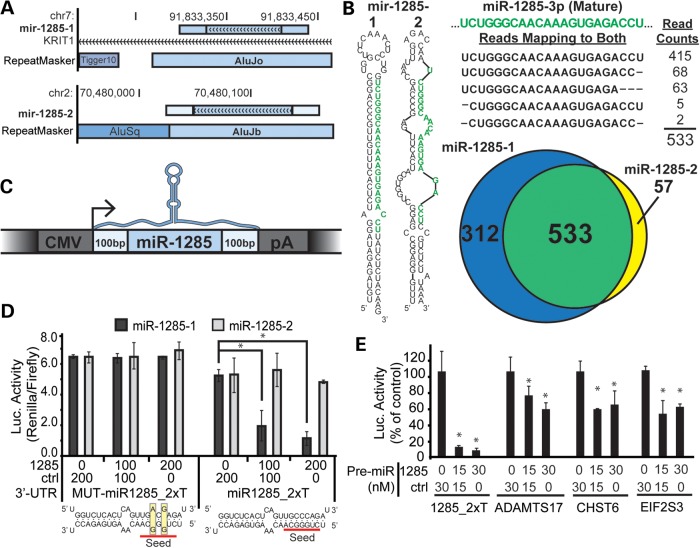
Alu-derived hsa-miR-1285-1 regulates homologous Alu-derived target sites. (A) Genome browser views of miR-1285-1 and miR-1285-2 loci are shown, including any overlapping transcript or RepeatMasker annotations. The miR-1285 precursor locus (light blue rectangle with arrows) is drawn to include the additional ∼200 flanking bases (light blue, no arrows). The miR-1285-1 isoform is located within an intron of the KRIT1 mRNA. (B) The most stable predicted secondary structure for human miR-1285-1 and miR-1285-2 precursors is reproduced from those provided in the miRBase repository. High-throughput sequencing reads mapping to the two loci were also taken from miRBase, and the reads mapping to the common mature miRNA sequence are shown, along with the total read counts. The mature miRNA sequence, as reported in miRBase, is highlighted in green. The Venn diagram summarizes the number of reads uniquely mapping to each locus. The area of the circles and region of overlap is drawn in proportion to the number of reads contained in each group. (C) A cartoon representation of the miR-1285 expression constructs is drawn. The coloring scheme is reproduced from Figure 5A. (D) Luciferase reporters expressing artificial 3′ UTR target sites for miR-1285 (miR1285_2xT), or a seed-mutant control (MUT-miR1285-2xT), were transfected into HEK293 cells along with the indicated doses of the miR-1285-1 (1285) and control (ctrl) expression plasmids (shown in A), and luciferase activity was measured 48 h later. Luciferase activity is represented as the measured ratio between Renilla and firefly luciferase. Predicted hybridizations between the miR-1285 mature sequence and the artificial target and mutant sites are shown below the corresponding luciferase data. (E) Luciferase reporters expressing the 3′ UTR of putative miR-1285 targets, ADAMTS17, CHST6 and EIF2S3, were co-transfected with the indicated concentrations of an artificial miR-1285 (1285) or control (ctrl) Pre-miR oligo. (C–E) N = 3 biological replicates with three technical replicates per transfection; error bars = SD. *P ≤ 0.05 (ANOVA; Tukey's post hoc).