Heparan sulfate (HS) and heparin are linear sulfated heteropolysaccharides consisting of alternating α1–4-linked D-glucosamine (GlcN) and 1–4-linked uronic acid (α-linkage for L-iduronic acid, IdoA, and β-linkage for D-glucuronic acid, GlcA). Possible modifications are 2-O-sulfation on the uronic acid residues and one or more modifications on the glucosamine residues including N-sulfation, N-acetylation, 6-O-sulfation, and 3-O-sulfation. Heparin and low molecular weight heparin (LMWH) are the most commonly used anticoagulants or antithrombotic drugs. Compared to HS, heparin has a higher level of sulfation and a higher IdoA content.[1] Heparin is mostly produced by mast cells and heparan sulfates are produced by different cell types in animals.[2] They are attractive synthetic targets due to the therapeutic application of heparin, and the important roles of HS and heparin in regulating cancer growth, blood coagulation, inflammation, assisting viral and bacterial infections, signal transduction, lipid metabolism, and cell differentiation.[3]
Synthetic heparins can eliminate the side effects caused by inherent heterogeneous heparins purified from natural sources. Their synthesis, however, possesses great synthetic challenges due to their structural complexity. Although much progress has been made over the last decade on the synthesis, analysis, and understanding of complex HS and heparin, the mechanisms for the formation and regulation of HS/heparin and the structure-function relationship of complex HS/heparin are still not fully understood.[2] A tailor-made synthetic process is still lacking.
Early chemical synthesis of heparin fragments and analogs[4] required many protection and deprotection steps, making the synthesis of even relatively small oligosaccharides time consuming and low efficient. More recently, various chemical synthetic approaches[5] including target-oriented,[6] modular,[7] combinatorial,[8] one-pot,[9] and solid-phase[10] syntheses, have been developed and used to produce HS/heparin oligosaccharides ranging from di- to octasaccharides of different sequences and sulfation patterns. Glycan microarrays have been developed to study heparin/HS-protein interactions.[11] Nevertheless, the synthesis of HS oligosaccharides of non-repetitive sequences is much more challenging than those with repetitive sequences. Synthetic efficiency decreases dramatically with the increase of the length of target molecules. Furthermore, the structure and length variations of targets can completely change the whole synthetic design.
Traditional methods of purifying oligosaccharides after enzymatic digestion of GAG chains have provided useful quantities of HS/heparin oligosaccharides for early structure–activity relationship studies.[12] Cloning and characterization of HS biosynthetic enzymes have allowed chemoenzymatic synthesis of various HS/heparin derivatives.[13] Bioactive HS structures that bind to antithrombin, fibroblast growth factor FGF2, or herpes simplex virus glycoprotein D (HSV gD) were enzymatically synthesized in milligram scales from completely desulfated and N-sulfated heparin by multiple O-sulfation using immobilized O-sulfotransferases with the regeneration of activated sulfate donor 3'-phosphoadenosine 5'-phosphosulfate (PAPS).[13c] Heparin-like polysaccharides with anticoagulant activity were obtained from purified Escherichia coli K5 capsular polysaccharide followed by C-5 epimerization of the uronic acid residues, chemical persulfation, and selective desulfonation.[13b] A heparin polysaccharide library with different sulfation patterns was obtained using different HS biosynthetic enzymes.[13d] Recently, two homogenous heparin oligosaccharides were chemoenzymatically synthesized in milligram scale and showed comparable activity as Arixtra (fondaparinux sodium), a synthetic anticoagulant heparin pentasaccharide.[14] Due to the complex nature of the HS biosynthesis and its involvement of multiple isoenzymes with overlappying substrate preferences, the exisiting methods do not provide precise control for the synthesis of many desired structures.
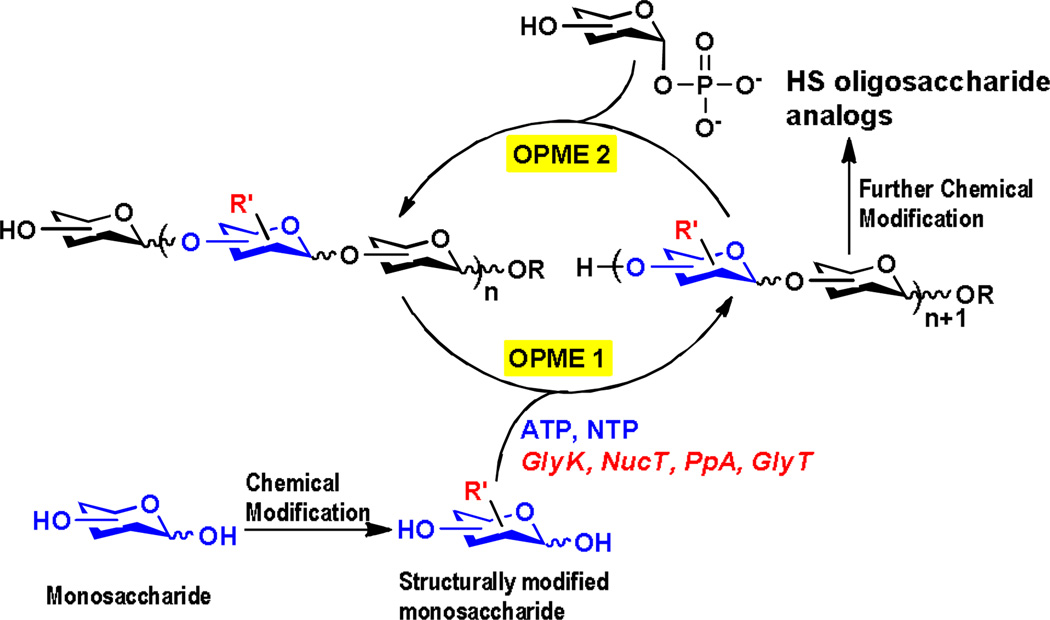
An efficient approach to synthesize carbohydrates containing post-glycosylational modifications (PGMs),[15] including HS oligosaccharides and analogs, with precise control is from the corresponding structurally modified monosaccharides using sequential one-pot multienzyme (OPME) glycosylation reactions (Scheme 1). The simplest OPME system consists of a monosaccharide kinase (GlyK), a nucleotidyltransferase (NucT), and a glycosyltransferase (GlyT) to allow the formation of desired glycosidic bonds in a regio- and stereo-specific manner. An inorganic pyrophosphatase (PpA) can also be used to remove the pyrophosphate by-product formed to drive the reaction towards the formation of sugar nucleotide. Further chemical modifications can be performed to introduce additional functionality after the formation of oligosaccharides. Here, we describe such a method for efficient synthesis of HS oligosaccharide analogs with desired sulfation patterns.
Scheme 1.
Sequential one-pot multienzyme (OPME) chemoenzymatic synthesis of heparan sulfate (HS) oligosaccharide analogs. GlyK, glycokinase; NucT, monosaccharide-1-phosphate nucleotidyltransferase; PpA, inorganic pyrophosphatase; GlyT, glycosyltransferase.
PmHS2 from Gram-negative bacterium Pasteurella multocida is a well studied heparosan synthase.[16] It is a bifunctional enzyme which has both α1–4-N-acetylglucosaminyltransferase (α1–4GlcNAcT) and β1–4-glucuronyltransferase (β1–4GlcAT) activities. PmHS2 cloned from Pasteurella multocida strain P-1059 (ATCC#15742, type A strain) has a good expression level in Escherichia coli (~17 mg/L cell culture). The α1–4GlcNAcT activity of PmHS2 is more promiscuous than that of Escherichia coli K5 α1–4GlcNAcT (KfiA) on using UDP-GlcNAc derivatives as donor substrates.[17]
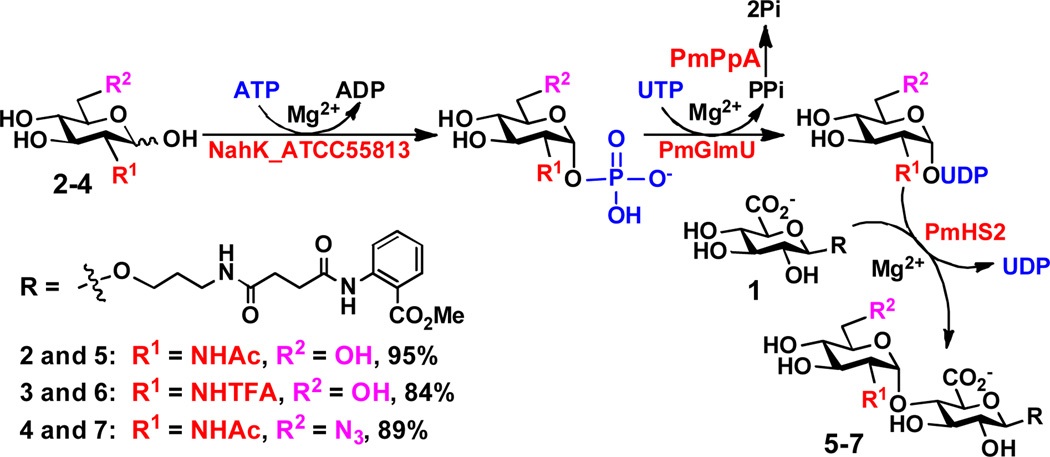
As shown in Scheme 2, using a fluorescently labeled glucuronide GlcAβ2AA (1) as the acceptor substrate and taking advantage of the substrate promiscuities of several enzymes involved in GlcNAc activativation and transfer, disaccharide GlcNAcα1–4GlcAβ2AA (5) and its derivatives GlcNTFAα1–4GlcAβ2AA (6) and GlcNAc6N3α1–4GlcAβ2AA (7) were synthesized in high yields (84–95%) from N-acetylglucosamine (GlcNAc, 2), N-trifluoroacetylglucosamine (GlcNTFA, 3), and 6-azido-6-deoxy-N-acetylglucosamine (GlcNAc6N3, 4), respectively, using a one-pot four-enzyme reaction containing NahK_ATCC55813,[18] PmGlmU,[19] PmPpA,[20] and PmHS2.[17] It was found that the cleavage of the N-TFA in 3 and 6 was significant at pH 7.5 but could be minimized by lowering the pH to 6.5 and shortening the reaction time.
Scheme 2.
One-pot four-enzyme GlcNAc activating and transfer system for the synthesis of GlcNAcα1–4GlcAβ2AA disaccharide and derivatives. Tris-HCl buffer (100 mM, pH 7.5) was used for the synthesis of disaccharides 5 and 7 and MES buffer (100 mM, pH 6.5) was used for the synthesis of disaccharide 6. NahK_ATCC55813, Bifidobacterium longum ATCC55813 N-acetylhexosamine 1-kinase; PmGlmU, Pasteurella multocida strain P-1059 (ATCC15742) N-acetylglucosamine-1-phosphate uridylyltransferase; PmPpA, Pasteurella multocida strain P-1059 inorganic pyrophosphatase; PmHS2, Pasteurella multocida strain P-1059 heparosan synthase 2.
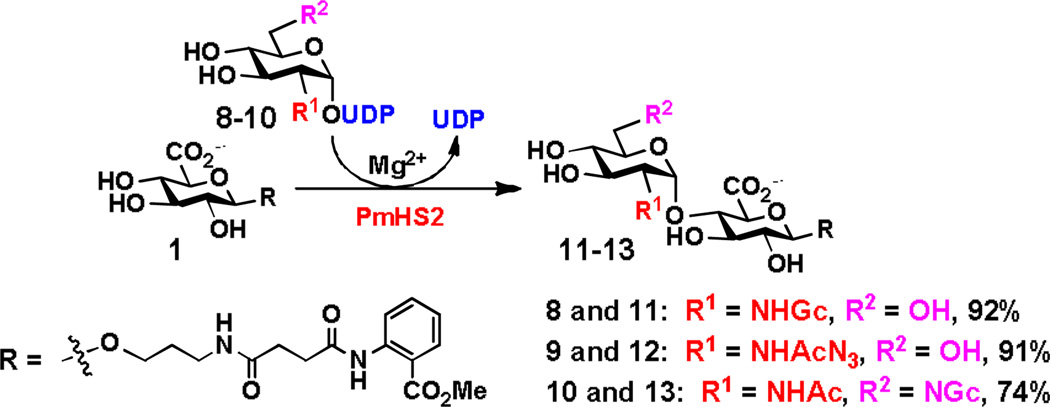
In addition, using three previously synthesized UDP-GlcNAc derivatives including UDP-N-glycolylglucosamine (UDP-GlcNGc, 8), UDP-N-azidoacetylglucosamine (UDP-GlcNAcN3, 9), and UDP-6-deoxy-6-glycolylamido-N-acetylglucosamine (UDP-GlcNAc6NGc, 10),[19] PmHS2-catalyzed GlcNAc transfer reaction produced disaccharides GlcNGcα1–4GlcAβ2AA (11), GlcNAcN3α1–4GlcAβ2AA (12), and GlcNAc6NGcα1–4GlcAβ2AA (13) in good to excellent yields (74–92%) (Scheme 3).
Scheme 3.
PmHS2-catalyzed synthesis of GlcNAcα1–4GlcAβ2AA disaccharide derivatives 11–13 in Tris-HCl buffer (100 mM, pH 7.5).
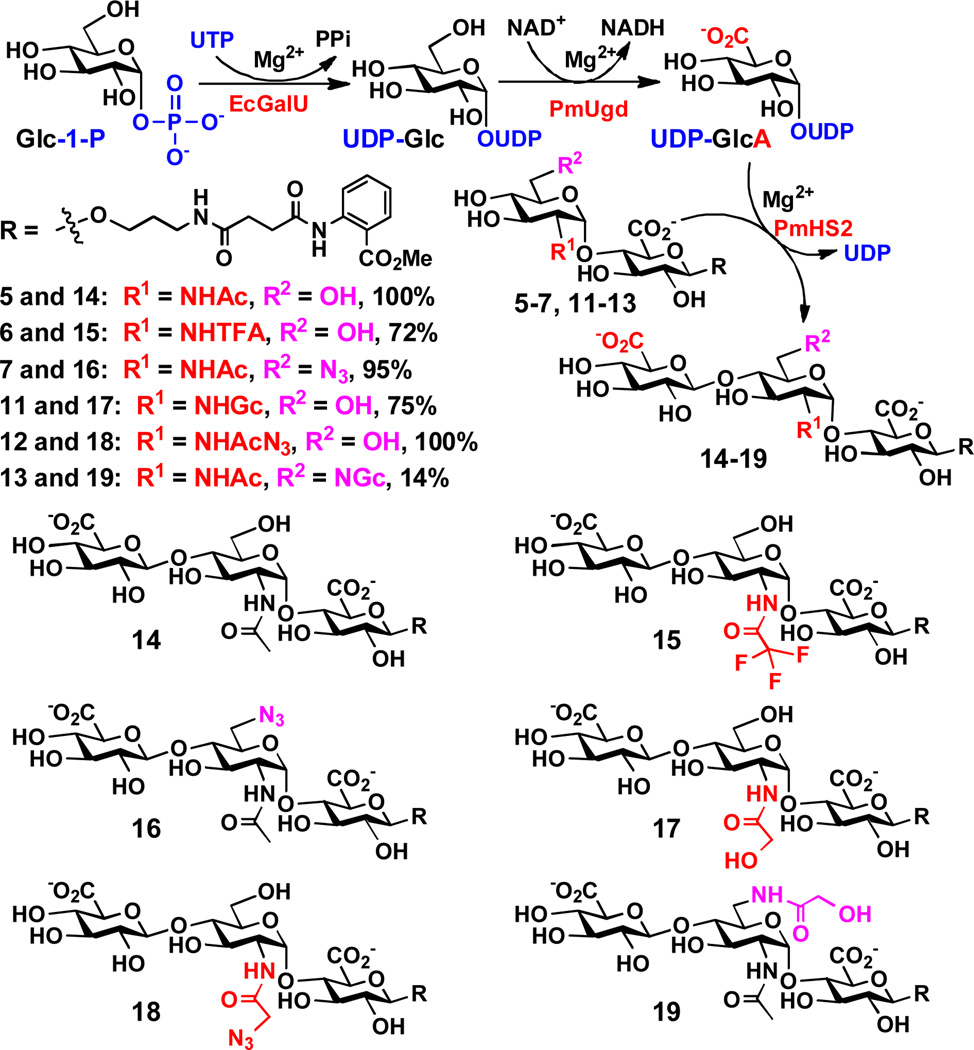
The obtained disaccharides 5–7 and 11–13 were used in a one-pot three-enzyme GlcA activation and transfer system (Scheme 4) to test the acceptor substrate specificity of the β1–4GlcAT activity of PmHS2. In this system, a glucose-1-phosphate uridylyltransferase cloned from Escherichia coli K-12 (EcGalU)[21] was used to catalyze the formation of UDP-Glc from Glc-1-P and UTP. A UDP-glucose dehydrogenase cloned from Pasteurella multocida strain P-1059 (PmUgd, see Supporting Information for cloning, purification, and DNA and protein sequences) was used in the presence of a coenzyme nicotinamide adenine dinucleotide (NAD+) to oxidize the C-6 of glucose (Glc) in UDP-Glc to form UDP-glucuronic acid (UDP-GlcA), which was used by the β1–4GlcAT activity of PmHS2 to form trisaccharides. The reactions were analyzed by a high-performance liquid chromatography (HPLC) system equipped with a fluorescent detector. As shown in Scheme 4, all disaccharides (5–7 and 11–13) tested were suitable acceptor substrates for the β1–4GlcAT activity of PmHS2. Trisaccharides GlcAβ1–4GlcNAcα1–4GlcAβ2AA (14), GlcAβ1–4GlcNAc6N3α1–4GlcAβ2AA (16), and GlcAβ1–4GlcNAcN3α1–4GlcAβ2AA (18) were formed in quantitative or near quantitative yields (95–100%). GlcAβ1–4GlcNTFAα1–4GlcAβ2AA (15) was formed in a relatively lower yield (72%), mainly due to the loss of the N-TFA-group to form another trisaccharide GlcAβ1–4GlcNH2α1–4GlcAβ2AA. Disaccharide GlcNGcα1–4GlcAβ2AA (11) with an N-glycolyl group at the C-2 of GlcN was a good acceptor for the β1–4GlcAT activity of PmHS2 to form GlcAβ1–4GlcNGcα1–4GlcAβ2AA (17) in 75% yield. In comparison, disaccharide GlcNAc6NGcα1–4GlcAβ2AA (13) with the same N-glycolyl group at the C-6 of GlcNAc was a poorer acceptor and trisaccharide GlcAβ1–4GlcNAc6NGcα1–4GlcAβ2AA (19) was formed in only 14% yield.
Scheme 4.
One-pot three-enzyme GlcA activation and transfer system for the synthesis of GlcAβ1–4GlcNAcα1–4GlcAβ2AA trisaccharide and derivatives. EcGalU, Escherichia coli K-12 glucose-1-phosphate uridylyltransferase; PmUgd, Pasteurella multocida strain P-1059 UDP-glucose dehydrogenase.
Taking together, these results indicate that in addition to the previously identified donor substrate promiscuity of the α1–4GlcNAcT activity of PmHS2,[17, 16b] the β1–4GlcAT activity of PmHS2 also has a broader acceptor substrate promiscuity than previously described.[22] These substrate promiscuities allowed the synthesis of GlcNAcα1–4GlcAβ2AA disaccharide derivatives containing modified GlcNAc residues which were further used for the formation of diverse trisaccharide derivatives. In principle, the same strategy can be repeated for the production of longer oligosaccharides and derivatives. The oligosaccharide derivatives can be used as important probes for investigating the acceptor binding pocket of PmHS2 which may bind to oligosaccharide acceptors longer than monosaccharides or disaccharides as discussed previously.[22]
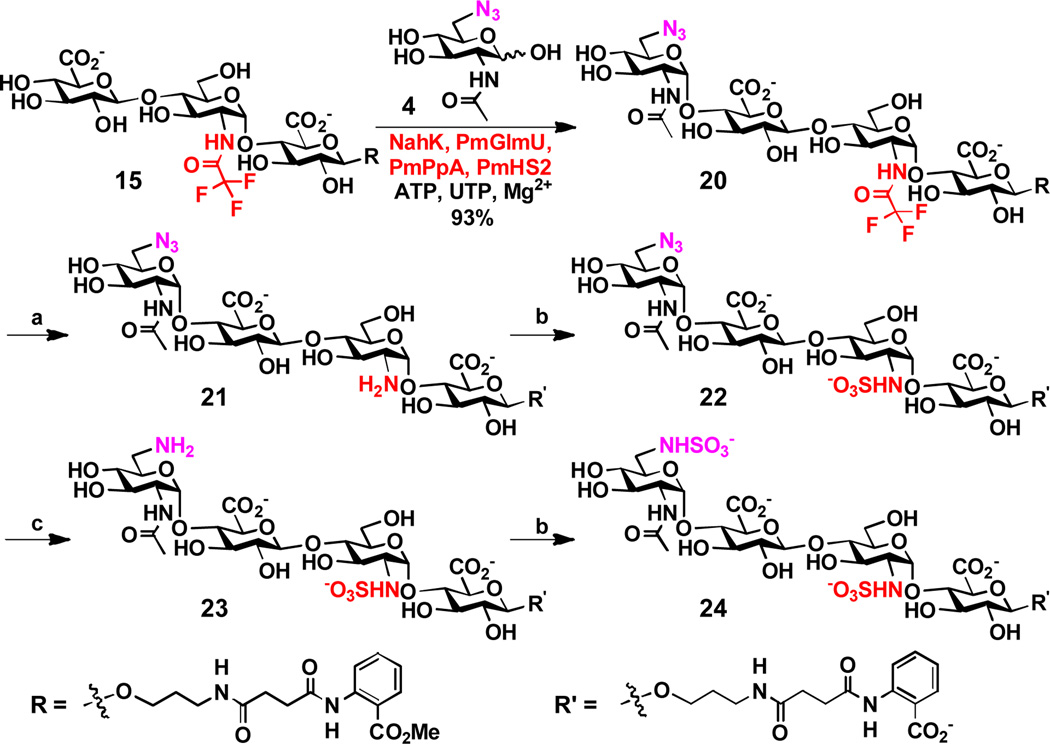
A strategy for preparative-scale synthesis of HS oligosaccharide analogs with controllable sulfation patterns was also explored. To do this, trisaccharide GlcAβ1–4GlcNTFAα1–4GlcAβ2AA (15) (>50 mg) was synthesized using the one-pot three-enzyme GlcA activation and transfer system shown in Scheme 4. The loss of the N-TFA group was minimized by carrying out the reaction at pH 7.0 instead of 7.5 and the yield of the reaction was increased to 87%. A shown in Scheme 5, the obtained trisaccharide 15 was used as a substrate in the one-pot four-enzyme GlcNAc activating and transfer system (the same to the one shown in Scheme 2) for the synthesis of tetrasaccharide GlcNAc6N3α1–4GlcAβ1–4GlcNTFAα1–4GlcAβ2AA (20) in high-yield (93%). The N-TFA group and the N3 group were individually converted to a free amine group as shown in tetrasaccharides 21 and 23, allowing sequential N-sulfation to generate HS tetrasaccharide analogs 22–24. More specifically, the N-TFA group at C-2 of the internal GlcNTFA residue in tetrasaccharide 20 was removed under mild basic conditions to produce tetrasaccharide GlcNAc6N3α1–4GlcAβ1–4GlcNH2α1–4GlcAβ2AA' (21) in 81% yield. As the removal of the N-TFA group was accompanied by the hydrolysis of the methyl carboxylate ester, tetrasaccharide 21 contains a free carboxyl group in the 2AA' motif instead of the carboxylic ester in the 2AA of tetrasaccharide 20. Sulfation of tetrasaccharide 21 for the formation of tetrasaccharide 22 required a larger excess of sulfation reagent (60 equiv.) and prolonged reaction time (3 days) to achieve a reasonable 70% yield. Catalytic hydrogenation of the azido group at the C-6 of the non-reduced end GlcNAc6N3 in tetrasaccharide 22 produced tetrasaccharide 23. Tetrasaccharide 24 was subsequently synthesized by N-sulfation.
Scheme 5.
One-pot four-enzyme synthesis of tetrasaccharide 20 and subsequent chemical derivatization reactions for the formation of tetrasaccharides 21–24. Reagents and conditions: (a) K2CO3, H2O, r.t. overnight, 81%; (b) complex Py•SO3, 2 M NaOH, H2O, 3 days, 70%; (c) H2, Pd/C, MeOH, H2O, 1 h, quantitative.
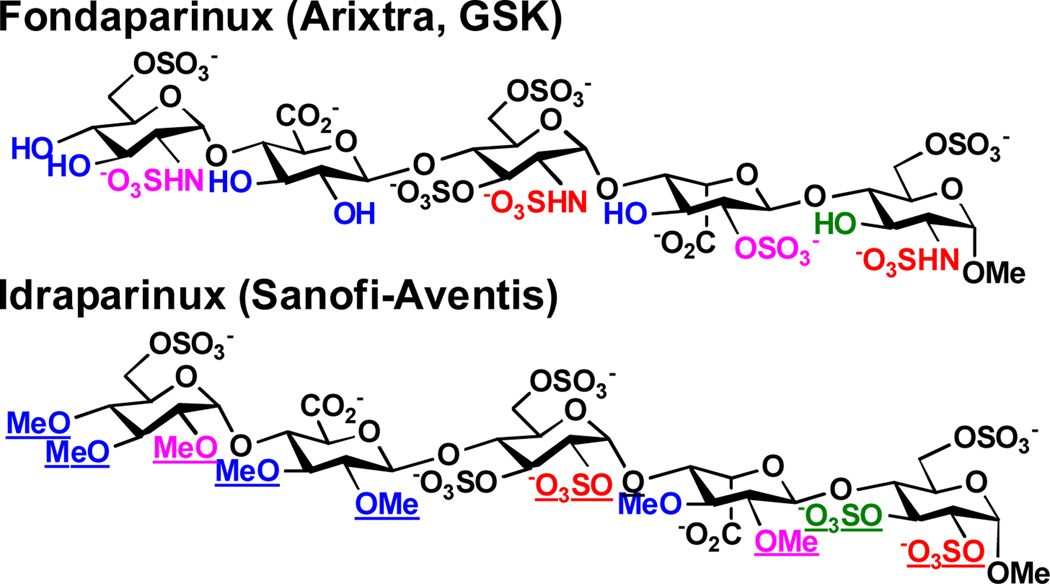
These N-sulfated oligosaccharide analogs mimic naturally occurring HS oligosaccharides and may have improved therapeutic potentials as they will most likely be more resistant to the activities of HS/heparin lyases and hydrolases. In deed, Idraparinux, a synthetic fully O-sulfated and O-methylated analog of heparin pentasaccharide, has higher anti-Xa activity and a longer duration of action compared to Fondaparinux (or Arixtra), an FDA-approved synthetic compound presenting a non-modified antithrombin-binding heparin pentasaccharide for the prevention and treatment of venous thromboembolism (Scheme 6).[23] The N-sulfated analogs of HS oligosaccharides may also have unexpected or improved functions.
Scheme 6.
Structural comparison of Fondaparinux (Arixtra) and Idraparinux. Differences are underlined.
In conclusion, we have developed efficient one-pot multienzyme (OPME) systems for GlcNAc and GlcA activation and transfer which have been used successfully for synthesizing N-sulfated analogs of HS oligosaccharides. We have demonstrated that PmHS2, a bifunctional heparosan synthase from Pasteurella multocida responsible for the production of N-acetylheparosan, has promiscuous donor and acceptor substrate activities. The method of synthesizing N-sulfated analogs of HS oligosaccharides in tailor-designed fashion will be explored further for synthesizing longer oligosaccharides and with more diverse sulfation patterns. The resulting compounds may compete efficiently with naturally occurring HS and heparin for binding to their target proteins and are potential therapeutics.
Experimental Section
One-pot four-enzyme synthesis of disaccharides 5–7
GlcAβ2AA 1 (5–30 mg, 1 eq.), GlcNAc or derivatives (1.5 eq.), ATP (1.8 eq.), and UTP (1.8 eq.) were dissolved in water in a 15 mL centrifuge tube containing MgCl2 (10 mM) in Tris-HCl (100 mM, pH 7.5) or MES (100 mM, pH 6.5) buffer. After adding NanK_ATCC55813 (0.5–2.1 mg), PmGlmU (1–3 mg), PmPpA (0.5–1.5 mg), and PmHS2 (1–6 mg), water was added to bring the concentration of GlcAβ2AA 1 to 5 mM. The reaction mixture was incubated for 12–36 h at 37 °C with gentle shaking.
PmHS2-catalyzed synthesis of disaccharides 11–13
GlcAβ2AA 1 (5–10 mg, 1 eq.) and UDP-GlcNAc derivatives (1.2 eq.) were dissolved in water in a 15 mL centrifuge tube containing MgCl2 (10 mM) in Tris-HCl buffer (100 mM, pH 7.5). After adding PmHS2 (1–2 mg), water was added to bring the volume of the reaction mixture to 10 mL. The reaction mixture was incubated for 12–36 h at 37 °C with gentle shaking. Product formation was monitored by thin-layer chromatography (EtOAc:MeOH:H2O = 4:2:1 by volume) with p-anisaldehyde sugar staining. The reaction was stopped by adding the same volume of ice-cold ethanol and incubating at 4 °C for 30 min. The mixture was concentrated and passed through a BioGel P-2 gel filtration column to obtain the desired product. Silica gel column purification (EtOAc:MeOH:H2O = 5:2:1) was applied when necessary to achieve further purification.
Small scale one-pot three-enzyme synthesis of trisaccharides 14–19
Typical reactions were performed in a total volume of 20 µL in Tris-HCl buffer (100 mM, pH 7.5) containing MgCl2 (10 mM), UTP (7.5 mM), disaccharides (5 mM), Glc-1-P (6 mM), NAD+ (12 mM), GalU (2.5 µg), PmUgd (8 µg), and PmHS2 (11.5 µg). Reactions were allowed to proceed for 12 h at 37 °C and quenched by adding ice-cold ethanol (20 µL) and water (1.96 mL) to make 100-fold dilution.
Supplementary Material
Footnotes
This work was partially supported by NSF grants CHE-0548235, CHE1012511, and NIH grant R01 HD065122. Bruker Avance-800 NMR spectrometer was funded by NSF grant DBIO-722538. X. Chen is a Camille Dreyfus Teacher-Scholar and a UC-Davis Chancellor’s Fellow.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Liu J, Pedersen L. Appl. Microbiol. Biotechnol. 2007;74:263–272. doi: 10.1007/s00253-006-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esko JD, Selleck SB. Ann. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 3.Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Ann. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 4.a) Sinay P, Jacquinet JC, Petitou M, Duchaussoy P, Lederman I, Choay J, Torri G. Carbohydr. Res. 1984;132:C5–C9. doi: 10.1016/s0008-6215(00)90633-5. [DOI] [PubMed] [Google Scholar]; b) Jacquinet JC, Petitou M, Duchaussoy P, Lederman I, Choay J, Torri G, Sinay P. Carbohydr. Res. 1984;130:221–241. doi: 10.1016/s0008-6215(00)90633-5. [DOI] [PubMed] [Google Scholar]
- 5.Noti C, Seeberger PH. Chem. Biol. 2005;12:731–756. doi: 10.1016/j.chembiol.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 6.a) Petitou M, van Boeckel CAA. Angew. Chem. Int. Ed. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]; b) Poletti L, Lay L. Eur. J. Org. Chem. 2003:2999–3024. [Google Scholar]
- 7.a) Lee JC, Lu XA, Kulkarni SS, Wen YS, Hung SC. J. Am. Chem. Soc. 2004;126:476–477. doi: 10.1021/ja038244h. [DOI] [PubMed] [Google Scholar]; b) Orgueira HA, Bartolozzi A, Schell P, Litjens RE, Palmacci ER, Seeberger PH. Chemistry. 2003;9:140–169. doi: 10.1002/chem.200390009. [DOI] [PubMed] [Google Scholar]; c) Werz DB, Seeberger PH. Chemistry. 2005;11:3194–3206. doi: 10.1002/chem.200500025. [DOI] [PubMed] [Google Scholar]; d) Chen C, Yu B. Bioorg. Med. Chem. Lett. 2009;19:3875–3879. doi: 10.1016/j.bmcl.2009.03.155. [DOI] [PubMed] [Google Scholar]; e) Tiruchinapally G, Yin Z, El-Dakdouki M, Wang Z, Huang X. Chemistry. 2011;17:10106–10112. doi: 10.1002/chem.201101108. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Zulueta MM, Lin SY, Lin YT, Huang CJ, Wang CC, Ku CC, Shi Z, Chyan CL, Irene D, Lim LH, Tsai TI, Hu YP, Arco SD, Wong CH, Hung SC. J. Am. Chem. Soc. 2012;134:8988–8995. doi: 10.1021/ja302640p. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Xu Y, Yang B, Tiruchinapally G, Sun B, Liu R, Dulaney S, Liu J, Huang X. Chemistry. 2010;16:8365–8375. doi: 10.1002/chem.201000987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arungundram S, Al-Mafraji K, Asong J, Leach FE, 3rd, Amster IJ, Venot A, Turnbull JE, Boons GJ. J. Am. Chem. Soc. 2009;131:17394–17405. doi: 10.1021/ja907358k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) DreefTromp CM, Willems HAM, Westerduin P, vanVeelen P, vanBoeckel CAA. Bioorg. Med. Chem. Lett. 1997;7:1175–1180. [Google Scholar]; b) Ojeda R, Terenti O, de Paz JL, Martin-Lomas M. Glycoconj. J. 2004;21:179–195. doi: 10.1023/B:GLYC.0000045091.18392.a8. [DOI] [PubMed] [Google Scholar]
- 11.a) de Paz JL, Noti C, Seeberger PH. J. Am. Chem. Soc. 2006;128:2766–2767. doi: 10.1021/ja057584v. [DOI] [PubMed] [Google Scholar]; b) Noti C, de Paz JL, Polito L, Seeberger PH. Chemistry. 2006;12:8664–8686. doi: 10.1002/chem.200601103. [DOI] [PubMed] [Google Scholar]
- 12.Hileman RE, Smith AE, Toida T, Linhardt RJ. Glycobiology. 1997;7:231–239. doi: 10.1093/glycob/7.2.231. [DOI] [PubMed] [Google Scholar]
- 13.a) Kuberan B, Beeler DL, Lech M, Wu ZL, Rosenberg RD. J. Biol. Chem. 2003;278:52613–52621. doi: 10.1074/jbc.M305029200. [DOI] [PubMed] [Google Scholar]; b) Lindahl U, Li JP, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T, Emeis J, Roberts I, Taylor C, Oreste P, Zoppetti G, Naggi A, Torri G, Casu B. J. Med. Chem. 2005;48:349–352. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]; c) Chen J, Avci FY, Munoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. J. Biol. Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Chen J, Jones CL, Liu J. Chem. Biol. 2007;14:986–993. doi: 10.1016/j.chembiol.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Zhang Z, McCallum SA, Xie J, Nieto L, Corzana F, Jimenez-Barbero J, Chen M, Liu J, Linhardt RJ. J. Am. Chem. Soc. 2008;130:12998–13007. doi: 10.1021/ja8026345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Masuko S, Takieddin M, Xu H, Liu R, Jing J, Mousa SA, Linhardt RJ, Liu J. Science. 2011;334:498–501. doi: 10.1126/science.1207478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H, Chen X. Org. Biomol. Chem. 2007;5:865–872. doi: 10.1039/b700034k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Chavaroche A, Springer J, Kooy F, Boeriu C, Eggink G. Appl. Microbiol. Biotechnol. 2010;85:1881–1891. doi: 10.1007/s00253-009-2214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sismey-Ragatz AE, Green DE, Otto NJ, Rejzek M, Field RA, DeAngelis PL. J. Biol. Chem. 2007;282:28321–28327. doi: 10.1074/jbc.M701599200. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Yu H, Thon V, Chen Y, Muthana MM, Qu J, Hie L, Chen X. Appl. Microbiol. Biotechnol. 2013 doi: 10.1007/s00253-013-4947-1. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Yu H, Chen Y, Lau K, Cai L, Cao H, Tiwari VK, Qu J, Thon V, Wang PG, Chen X. Molecules. 2011;16:6396–6407. doi: 10.3390/molecules16086396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Thon V, Li Y, Yu H, Ding L, Lau K, Qu J, Hie L, Chen X. Chem. Commun. 2011;47:10815–10817. doi: 10.1039/c1cc14034e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau K, Thon V, Yu H, Ding L, Chen Y, Muthana MM, Wong D, Huang R, Chen X. Chem. Commun. 2010;46:6066–6068. doi: 10.1039/c0cc01381a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Fang JW, Zhang JB, Liu ZY, Shao J, Kowal P, Andreana P, Wang PG. J. Am. Chem. Soc. 2001;123:2081–2082. doi: 10.1021/ja005738v. [DOI] [PubMed] [Google Scholar]
- 22.Otto NJ, Green DE, Masuko S, Mayer A, Tanner ME, Linhardt RJ, DeAngelis PL. J. Biol. Chem. 2012;287:7203–7212. doi: 10.1074/jbc.M111.311704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harenberg J. Thromb. Haemost. 2009;102:811–815. doi: 10.1160/TH09-08-0555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.