Figure 1.
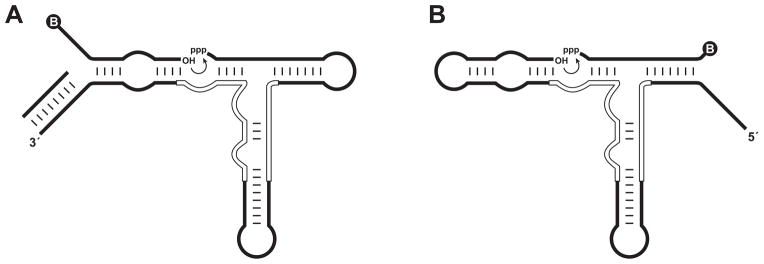
Formats for in vitro evolution of RNA ligation activity. (A) Substrate-A format, employing a separate 5′-biotinylated molecule bearing a 3′-hydroxyl. The primer for reverse transcription is pre-hybridized to the 3′ end of the enzyme to prevent interaction of this region with the substrate. (B) Substrate-B format, employing a separate 3′-biotinylated molecule bearing a 5′-triphosphate. Curved arrow indicates the site of ligation. Circled “B” indicates the biotin moiety. Open lines within the enzyme indicate nucleotides that were mutagenized at 21% degeneracy in the starting population.
