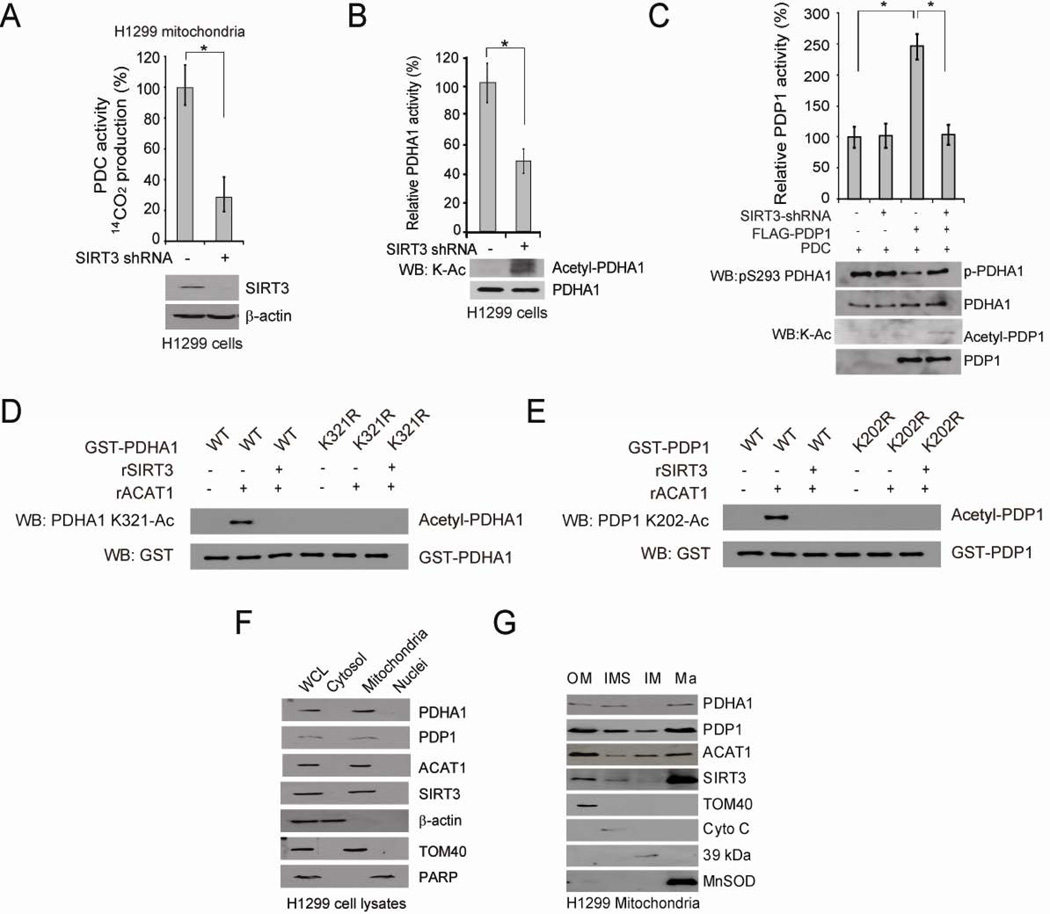
Figure 5. Mitochondrial SIRT3 is the upstream deacetylase of PDHA1 and PDP1.
(A) Control vector and stable SIRT3 knockdown cells were tested for PDC flux (upper). SIRT3 protein level was detected by Western blot (lower).
(B–C) Purified PDHA1 (B) and PDP1 (C) were incubated with cell lysates from H1299 cells with stable knockdown of SIRT3 or an empty vector, followed by PDHA1 and PDP1 activity assays (upper panels) and Western blot (lower panels).
(D–E) Purified PDHA1 WT and K321R mutant (D) or PDP1 WT and K202R mutant (E) were treated with rACAT1, followed by incubation with recombinant SIRT3 and Western blot.
(F) Western blot results show co-localization of PDHA1, PDP1, ACAT1 and SIRT3 in mitochondria of H1299 cells. Cytosolic β-actin, mitochondrial TOM 40 and nuclear PARP were included as control markers.
(G) Western blot results show co-localization of PDHA1, PDP1, ACAT1 and SIRT3 in the outer membrane (Om), inter-membrane space (IMS) and matrix (Ma) of mitochondria in H1299 cells. TOM40, Cyto C, complex I 39-kDa protein, and MnSOD are markers for Om, IMS, inner membrane (Im), and Ma, respectively. The error bars represent mean values +/− SD.
Also see Figure S5.