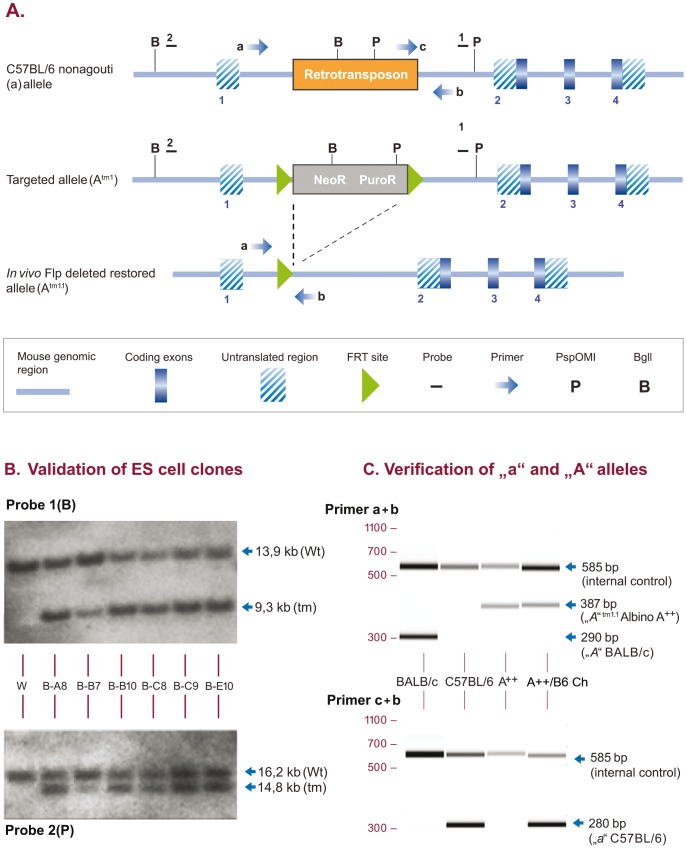
Figure 1. Restoration of the agouti locus in C57BL/6NTac ES cells and mice.
(A) Gene targeting strategy. The Neomycin (NeoR)/Puromycin (PuroR) selection marker is displayed as a grey box (neoR and puroR). Numbered dashes indicate external probes used for Southern Blot analysis of ES cell clones, lettered arrows indicate oligos used for genotyping of mice. Deletion of the FRT flanked selection was obtained in vivo simultaneously with germline transmission. (B) Southern Blot validation with genomic DNA isolated from 6 ES cell clones and wildtype C57BL/6 genomic DNA as a control (W) using external probe 1 in combination with BglI restriction digest (upper panel, Probe 1, B) leading to a wildtype allele of 13,9 kb (Wt) and a targeted allele of 9,3 kb (tm) and confirmatory Southern Blot validation with external probe 2 in combination with PspOMI restriction digest (lower panel, Probe 2, P), leading to a wildtype allele of 16,2 kb (Wt) and a targeted allele of 14,8 kb (tm). (C) PCR verification. Clone B-B10 was selected for chimera generation and germline transmission. Primer combinations a + b (upper panel) resulted in amplification of a 290 bp wildtype A allele in BALB/c control mice (lane 1, BALB/c) and a 387 bp restored Atm1.1 allele in homozygous Albino A++ mice (lane 3,A++) and in chimeras generated with C57BL/6 ES cells injected in homozyogus A++ host embryos (lane 4, A++/B6 Ch). Primer combinations c + b (lower panel) amplified a 280 bp C57BL/6 wildtype a allele in C57BL/6 control mice (lane 2, C57BL/6) and in A++/B6 Ch (lane 4, A++/B6 Ch). Note: PCR amplicons are of different size for the BALB/c A and the A++ restored Atm1.1 alleles. Also, in contrast to homozygous A++ mice, A++/B6 Ch amplify both, the ES cell derived a and the A++ derived Atm1.1 alleles.