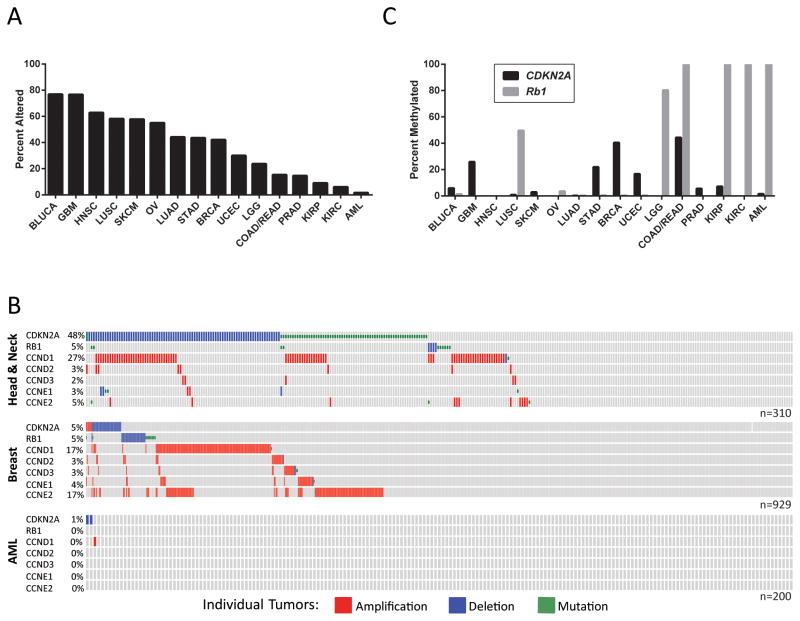
Figure 4.
Alterations in the p16INK4a Tumor Suppressor Pathway are Frequent in Human Cancer. A, Data from TCGA was obtained and analyzed using cBioPortal (112, 113). The tumors analyzed are as follows: Bladder Urothelial Carcinoma (BLUCA), Glioblastoma Multiforme (GBM), Head and Neck Squamous Cell Carcinoma (HNSC), Lung Squamous Cell Carcinoma (LUSC), Skin Cutaneous Melanoma (SKCM), Ovarian Serous Cystadenocarcinoma (OV), Lung Adenocarcinoma (LUAD), Stomach Adenocarcinoma (STAD), Breast Invasive Cancer (BRCA), Uterine Corpus Endometrial Carcinoma (UCEC), Brain Lower Grade Glioma (LGG), Colon and Rectum Adenocarcinoma (COAD/READ), Prostate Adenocarcinoma (PRAD), Kidney Renal Papillary Cell Carcinoma (KIRP), Kidney Renal Clear Cell Carcinoma (KIRC), Acute Myeloid Leukemia (AML). The percent of tumors with mutations or copy number changes in the p16INK4a tumor suppressor pathway are shown. For the purposes of this analysis the p16INK4a tumor suppressor pathway was defined to contain: CDKN2A (p16INK4a), RB1 (RB), CCND1 (CYCLIN D1), CCND2 (CYCLIN D2), CCND3 (CYCLIN D3), CCNE1 (CYCLIN E1), and CCNE2 (CYCLIN E2). B, OncoPrints from cBioPortal show aberrations in individual tumors across the X-axis. C, Methylation of CDKN2A (black bars) and RB1 (grey bars) was quantified using HM27 or HM450 TCGA data. Genes were considered methylated if β-values exceeded 0.2.