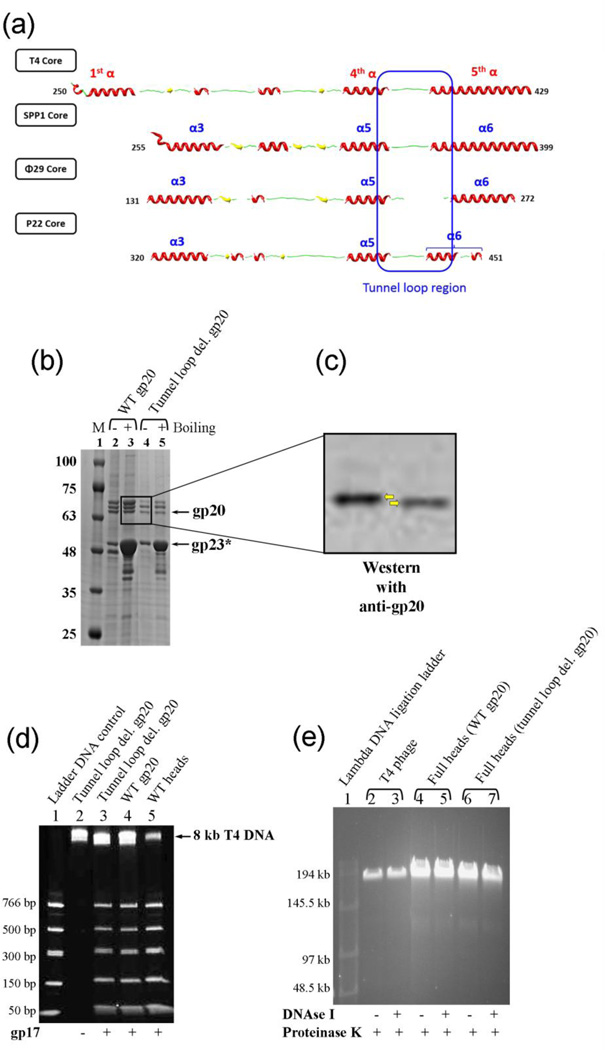
Fig. 9.
The tunnel loops are not essential for DNA translocation. (a) Secondary structure alignment of the tunnel loop regions of the portal proteins from different phages. The solved structures of different portal proteins are aligned with the predicted secondary structure of the T4 gp20 homology model f (Fig. 2). The presence of the tunnel loop between the inner helix α4 of portal channel and the kinked helix α5 of wing domain is a conserved feature among all the phage portals. (b) 4–20% gradient (w/v) SDS-PAGE showing the unboiled (−) and 5-min boiled (+) samples of the tunnel loop deletion head particles. M represents the molecular weight standards. The positions of gp20 and gp23* bands are marked with arrows. The gp20 band of the deletion mutant (lanes 4 and 5) migrated slightly faster than the WT gp20 (lanes 2 and 3). (c) The same gel was subjected to Western blotting using gp20 antiserum to confirm the positions of gp20 bands (indicated by the yellow arrows). (d) 4–20% PAGE showing the DNA packaging results of the tunnel loop deletion mutant heads. Approximately 2 × 1010 head particles and 2 µM gp17 were used to package the 50–766 bp ladder DNA. Heads assembled with WT gp20 and WT heads prepared from phage-produced portal were used as positive controls (lanes 4 and 5). The negative control lacked gp17 (lane 2). The position of the 8 kb T4 DNA retained in the heads is indicated. (e) T4 phage (5 × 108 particles per lane; lanes 2 and 3) or full heads (2 × 109 particles per lane; lanes 4–7) were treated with DNAse I (37°C, 30 min) followed by proteinase K (65°C, 30 min) or proteinase K only, as shown by “+” or “−” under the figure, and subjected to pulse field (field inversion) agarose gel (0.8% w/v) electrophoresis. The phage λ DNA ligation ladder (lane 1) was used to determine the size of DNA present in phage or heads.